- Optimized protocol on the IP-Star Compact for reproducible ChIP-qPCR results
- Shorter protocol - DNA purification module takes only 30 minutes reducing time from the standard 5 hours
- Ideal for histone and non-histone proteins – Use 1 million cells for histone marks or 4 million cells for transcription factors. The protocol allows the use of fewer cells per IP. The minimum number of cells will depend on the abundance of the protein in your sample.
- Suitable for cells and for tissues samples – For tissues, use 7 mg per IP for histone marks or 30 mg per IP for transcription factors. The protocol allows the use of less tissue per IP. The minimum amount of tissue will depend on the abundance of the protein in your sample.
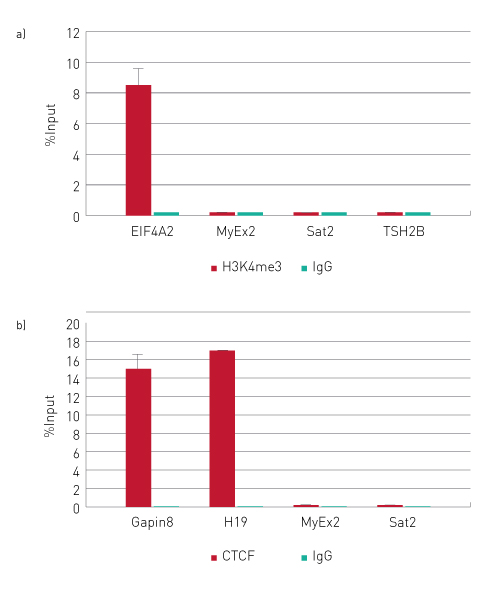
Chromatin immunoprecipitation analysis using control H3K4me3 and CTCF antibodies:
ChIP was performed on IP-Star Compact Automated System using the Auto iDeal ChIP-qPCR Kit, the antibodies (A) anti-H3K4me3 (Diagenode, Cat. no. C15410003) and (B) anti-CTCF (Diagenode, Cat. no. C15410210) . Sheared chromatin from 1 million (A) and 4 million (B) cells human HeLa cells, 1 µg of the positive control antibody and 1 µg of the negative IgG control (Diagenode, Cat. no. C15410206) were used per IP. The qPCR reactions were performed using primers for specific positive and negative loci. The figure shows the recovery expressed as a percentage of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). Error bars represent the standard deviation of at least 2 biological repetitions .
