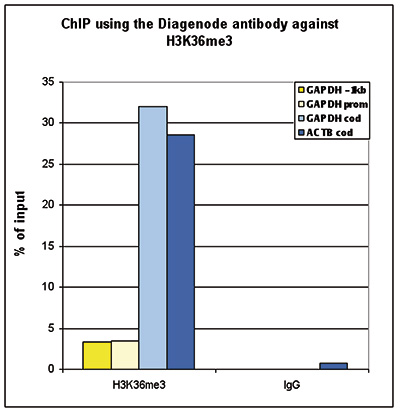
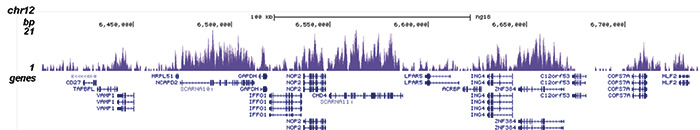
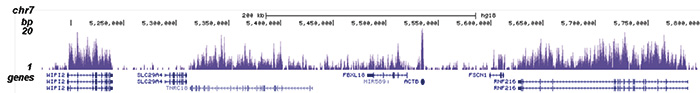
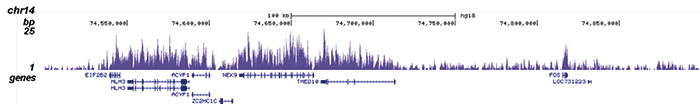
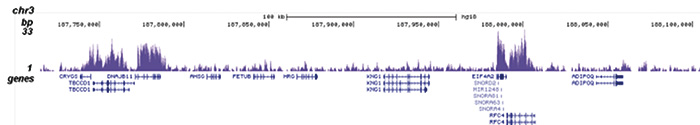
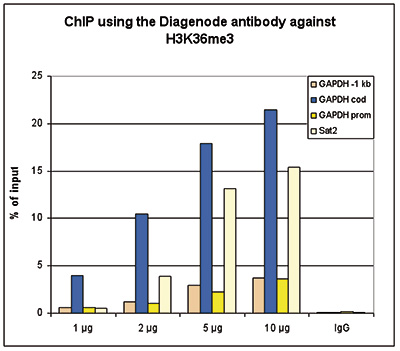
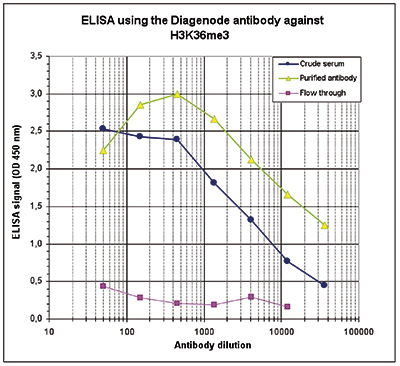
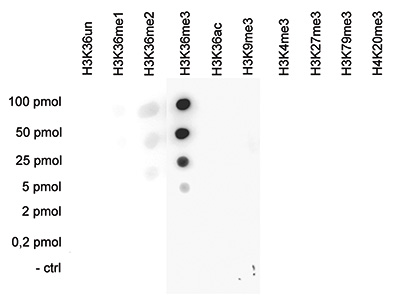
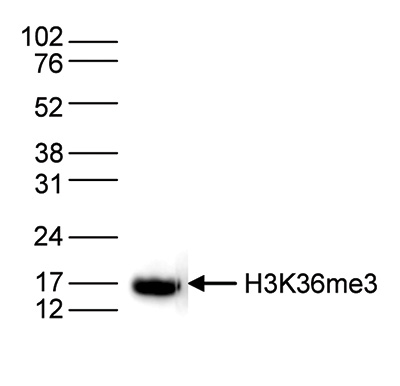
How to properly cite our product/service in your work We strongly recommend using this: H3K36me3 polyclonal antibody (sample size) (Hologic Diagenode Cat# C15410058-10 Lot# A8889-001P). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Multimodal epigenetic and enhancer network remodeling shape the transcriptional landscape of human beige adipocytes
Hazell Pickering, Sarah et al.
Epigenetic regulation is a key determinant of adipocyte fate, driving the differentiation toward white or thermogenic beige phenotypes in response to environmental cues. To dissect the mechanisms orchestrating this plasticity in human adipocytes, we conducted an integrative analysis of transcriptomic, epigenomic... |
Multimodal epigenetic and enhancer network remodeling shape the transcriptional landscape of human beige adipocytes
Hazell Pickering, Sarah et al.
Abstract
Epigenetic regulation is a key determinant of adipocyte fate, driving the differentiation toward white or thermogenic beige phenotypes in response to environmental cues. To dissect the mechanisms orchestrating this plasticity in human adipocytes, we conducted an integrative analysis of transcriptomic, epig... |
Chromatin profiling identifies transcriptional readthrough as a conservedmechanism for piRNA biogenesis in mosquitoes.
Qu J. et al.
The piRNA pathway in mosquitoes differs substantially from other model organisms, with an expanded PIWI gene family and functions in antiviral defense. Here, we define core piRNA clusters as genomic loci that show ubiquitous piRNA expression in both somatic and germline tissues. These core piRNA clusters are enriche... |
Gene Regulatory Interactions at Lamina-Associated Domains
Madsen-Østerbye J. et al.
The nuclear lamina provides a repressive chromatin environment at the nuclear periphery. However, whereas most genes in lamina-associated domains (LADs) are inactive, over ten percent reside in local euchromatic contexts and are expressed. How these genes are regulated and whether they are able to interact with regu... |
Epigenetic Mechanisms Mediating Cell State Transitions in Chondrocytes
Wuelling M. et al.
Epigenetic modifications play critical roles in regulating cell lineage differentiation, but the epigenetic mechanisms guiding specific differentiation steps within a cell lineage have rarely been investigated. To decipher such mechanisms, we used the defined transition from proliferating (PC) into hypertrophic chon... |
p53 directly represses human LINE1 transposons.
Tiwari, Bhavana and Jones, Amanda E and Caillet, Candace J and Das, Simantiand Royer, Stephanie K and Abrams, John M
p53 is a potent tumor suppressor and commonly mutated in human cancers. Recently, we demonstrated that p53 genes act to restrict retrotransposons in germline tissues of flies and fish but whether this activity is conserved in somatic human cells is not known. Here we show that p53 constitutively restrains human LINE... |
Premature termination codons in the gene cause reduced local mRNA synthesis.
García-Rodríguez R, Hiller M, Jiménez-Gracia L, van der Pal Z, Balog J, Adamzek K, Aartsma-Rus A, Spitali P
Duchenne muscular dystrophy (DMD) is caused by mutations in the gene leading to the presence of premature termination codons (PTC). Previous transcriptional studies have shown reduced DMD transcript levels in DMD patient and animal model muscles when PTC are present. Nonsense-mediated decay (NMD) has been suggested ... |
The Elongin Complex Antagonizes the Chromatin Factor Corto for Vein versus Intervein Cell Identity in Drosophila Wings.
Rougeot J, Renard M, Randsholt NB, Peronnet F, Mouchel-Vielh E
Drosophila wings mainly consist of two cell types, vein and intervein cells. Acquisition of either fate depends on specific expression of genes that are controlled by several signaling pathways. The nuclear mechanisms that translate signaling into regulation of gene expression are not completely understood, but they... |
Differential transcript isoform usage pre- and post-zygotic genome activation in zebrafish.
Aanes H, Østrup O, Andersen IS, Moen LF, Mathavan S, Collas P, Alestrom P
BACKGROUND: Zebrafish embryos are transcriptionally silent until activation of the zygotic genome during the 10th cell cycle. Onset of transcription is followed by cellular and morphological changes involving cell speciation and gastrulation. Previous genome-wide surveys of transcriptional changes only assessed gene... |
Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer.
Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, Li G, Mittler G, Liu ET, Bühler M, Margueron R, Schneider R
Histone modifications are key regulators of chromatin function. However, little is known to what extent histone modifications can directly impact on chromatin. Here, we address how a modification within the globular domain of histones regulates chromatin function. We demonstrate that H3K122ac can be sufficient to st... |
Prepatterning of developmental gene expression by modified histones before zygotic genome activation.
Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Østrup O, Winata C, Mathavan S, Müller F, Aleström P, Collas P
A hallmark of anamniote vertebrate development is a window of embryonic transcription-independent cell divisions before onset of zygotic genome activation (ZGA). Chromatin determinants of ZGA are unexplored; however, marking of developmental genes by modified histones in sperm suggests a predictive role of histone m... |
Characterization of the contradictory chromatin signatures at the 3' exons of zinc finger genes.
Blahnik KR, Dou L, Echipare L, Iyengar S, O'Geen H, Sanchez E, Zhao Y, Marra MA, Hirst M, Costello JF, Korf I, Farnham PJ
The H3K9me3 histone modification is often found at promoter regions, where it functions to repress transcription. However, we have previously shown that 3' exons of zinc finger genes (ZNFs) are marked by high levels of H3K9me3. We have now further investigated this unusual location for H3K9me3 in ZNF genes. Neither ... |
Promoter-exon relationship of H3 lysine 9, 27, 36 and 79 methylation on pluripotency-associated genes.
Barrand S, Andersen IS, Collas P
Evidence links pluripotency to a gene regulatory network organized by the transcription factors Oct4, Nanog and Sox2. Expression of these genes is controlled by epigenetic modifications on regulatory regions. However, little is known on profiles of trimethylated H3 lysine residues on coding regions of these genes in... |
Chromatin states of core pluripotency-associated genes in pluripotent, multipotent and differentiated cells.
Barrand S, Collas P
Oct4, Nanog and Sox2 constitute a core of transcription factors controlling pluripotency. Differentiation and reprogramming studies have unraveled a few epigenetic modifications associated in relation to the expression state of OCT4, NANOG and SOX2. There is, however, no comprehensive map of chromatin states on thes... |