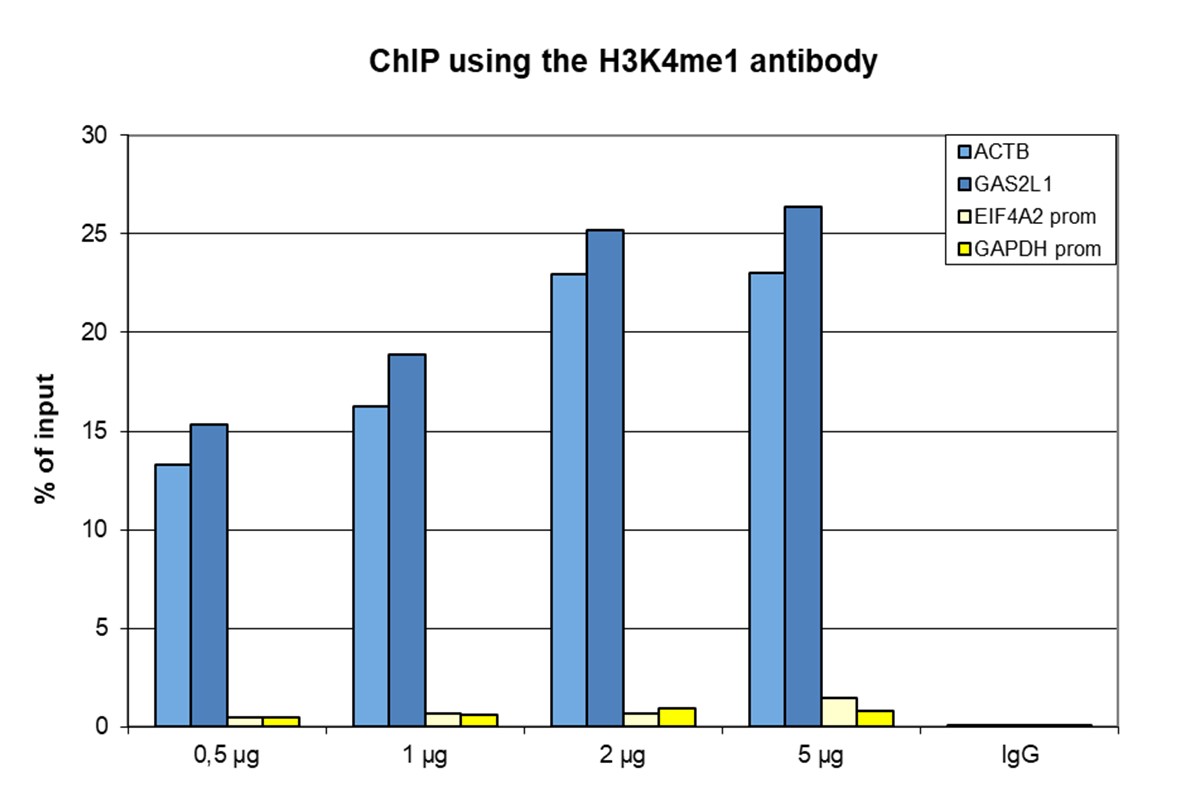
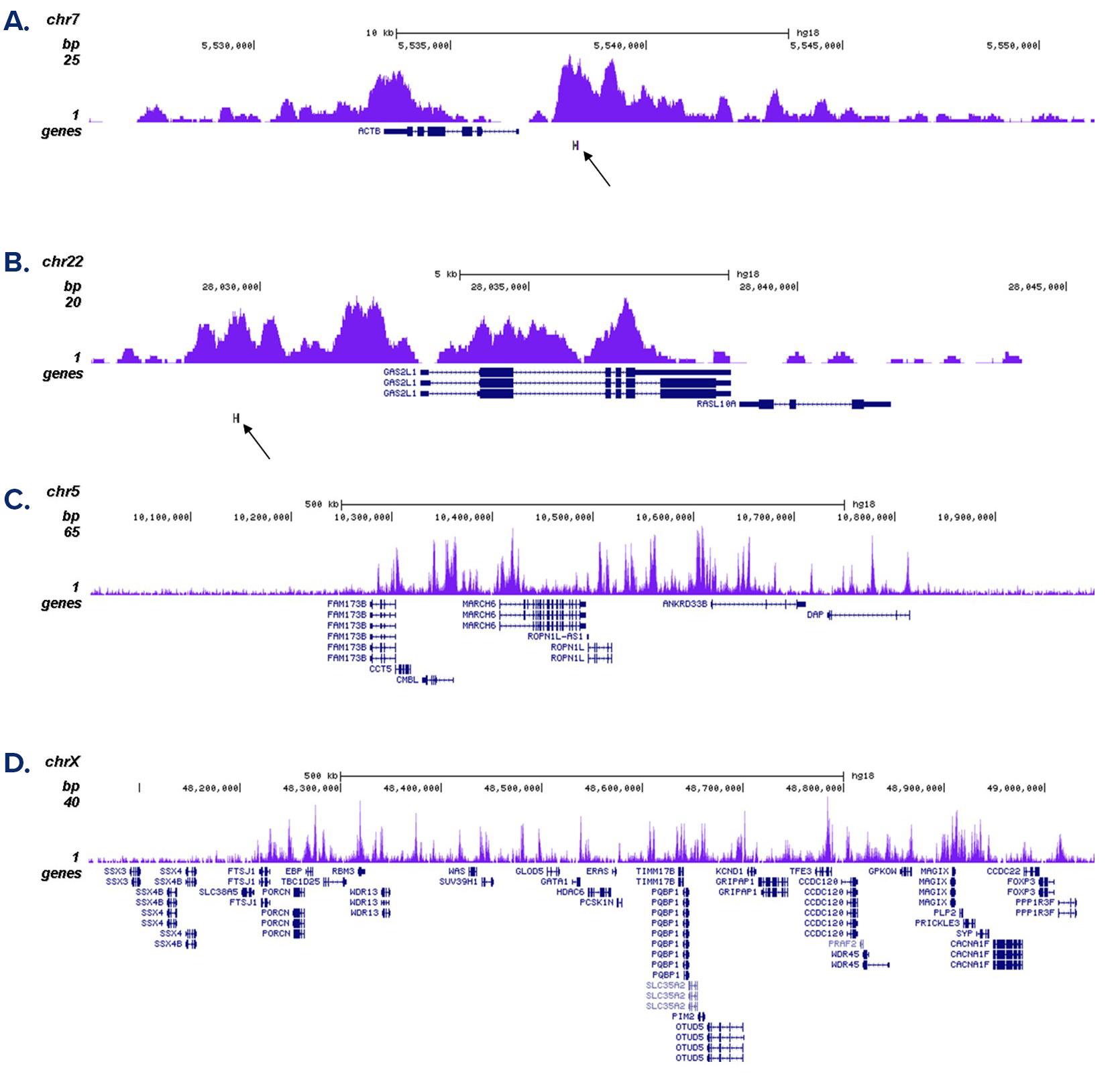
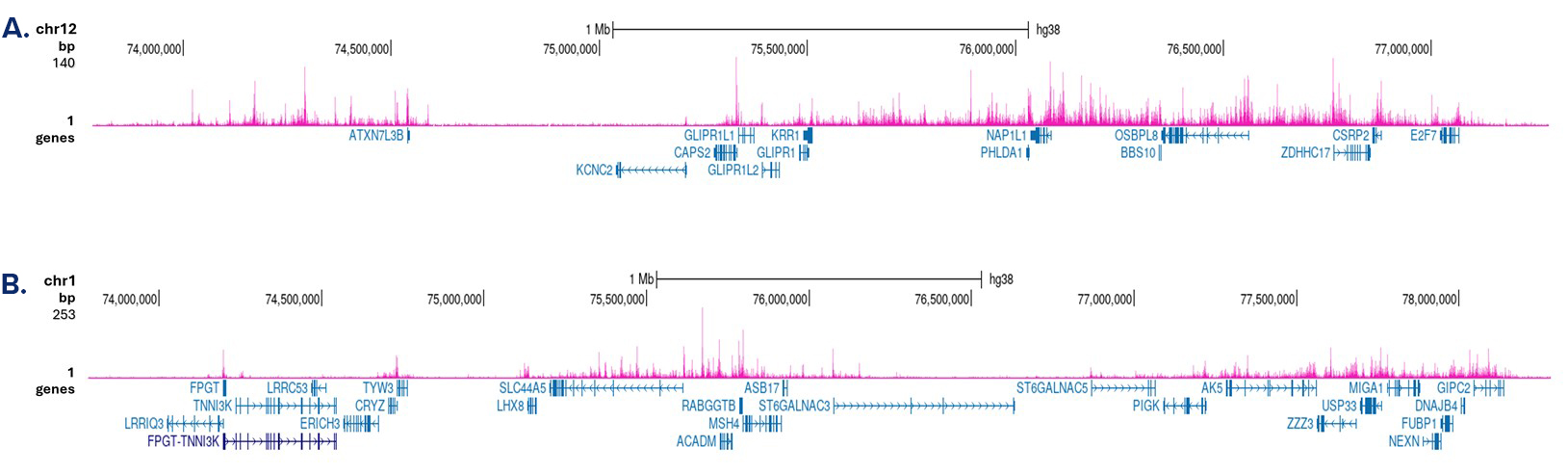
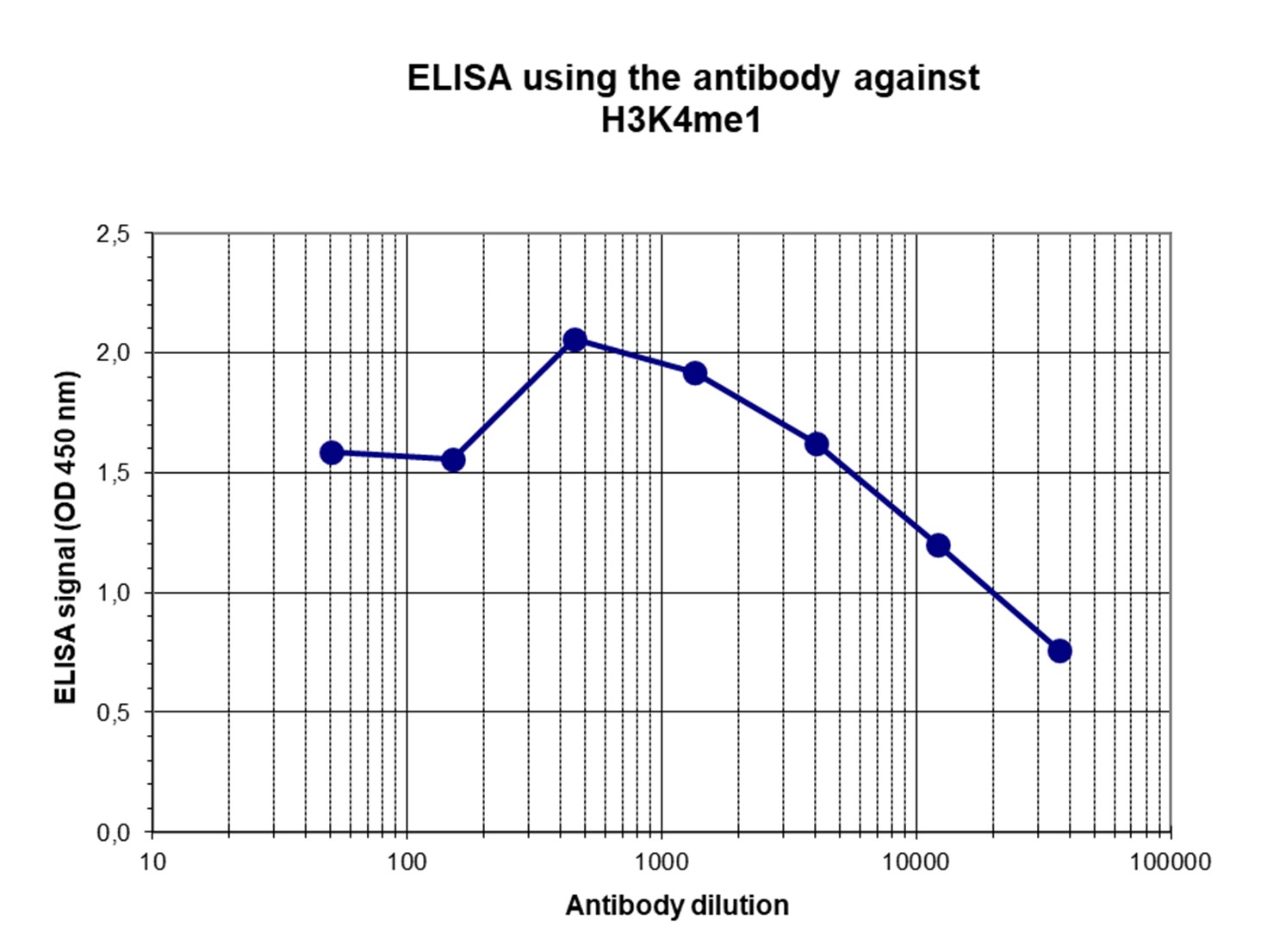
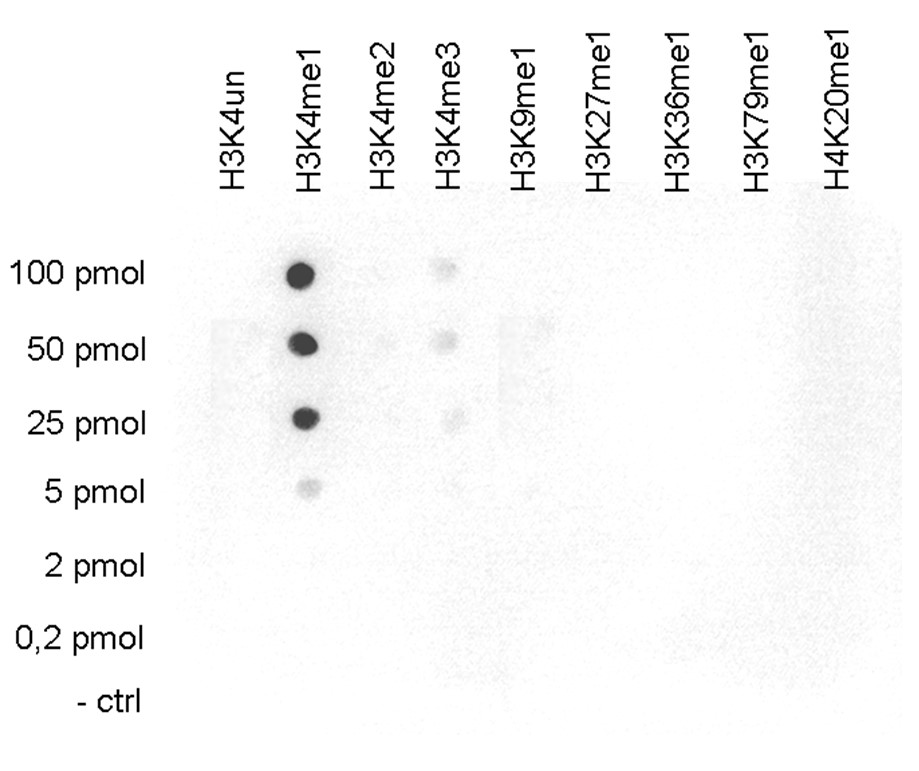
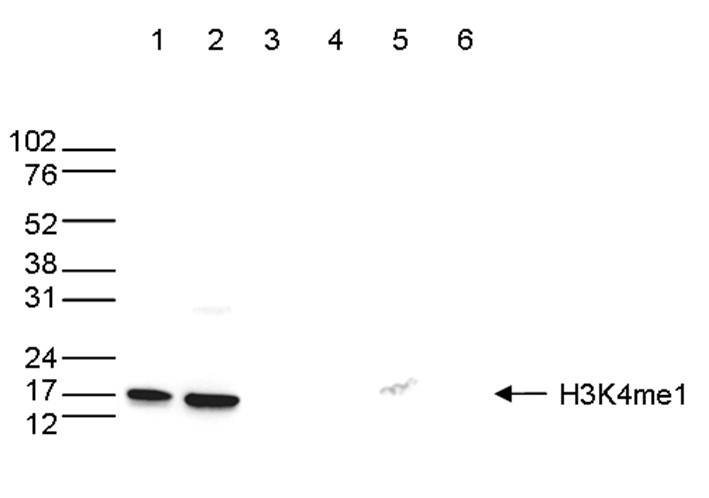
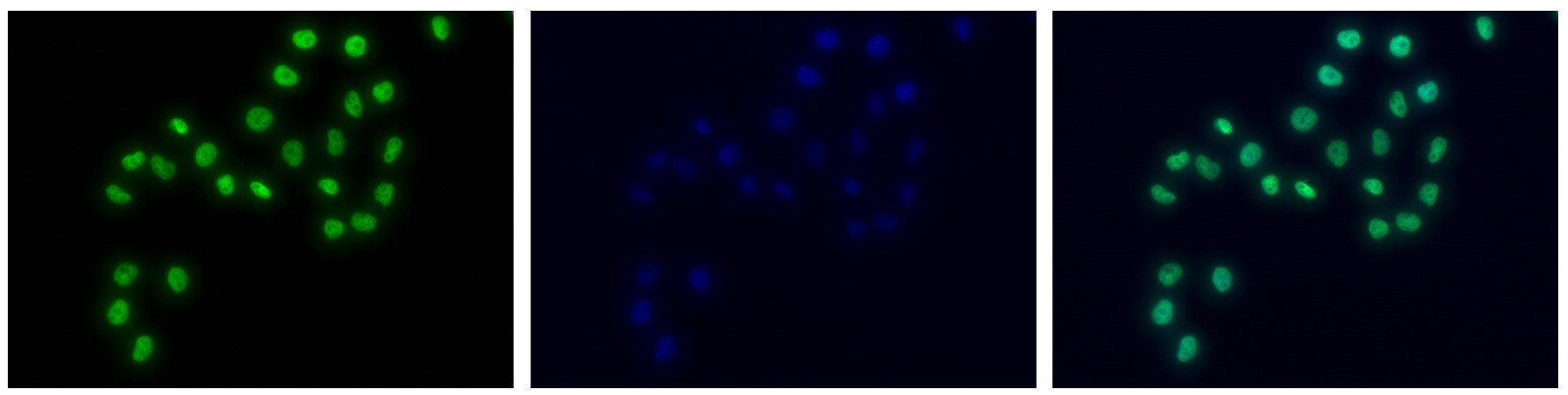
How to properly cite our product/service in your work We strongly recommend using this: H3K4me1 polyclonal antibody (sample size) (Hologic Diagenode Cat# C15410037-10 Lot# A1657D). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Multimodal epigenetic and enhancer network remodeling shape the transcriptional landscape of human beige adipocytes
Hazell Pickering, Sarah et al.
Epigenetic regulation is a key determinant of adipocyte fate, driving the differentiation toward white or thermogenic beige phenotypes in response to environmental cues. To dissect the mechanisms orchestrating this plasticity in human adipocytes, we conducted an integrative analysis of transcriptomic, epigenomic... |
Multimodal epigenetic and enhancer network remodeling shape the transcriptional landscape of human beige adipocytes
Hazell Pickering, Sarah et al.
Abstract
Epigenetic regulation is a key determinant of adipocyte fate, driving the differentiation toward white or thermogenic beige phenotypes in response to environmental cues. To dissect the mechanisms orchestrating this plasticity in human adipocytes, we conducted an integrative analysis of transcriptomic, epig... |
Extrusion fountains are restricted by WAPL-dependent cohesin release and CTCF barriers
Liu, Ning Qing et al.
Interphase chromosomes are mainly shaped by loop extrusion and compartmentalisation mechanisms. However, their temporal component and cause-effect relationships remain largely unknown. In this study, we use acute degradation of WAPL, CTCF and cohesin in mouse embryonic stem cells to investigate the dynamics of l... |
Single-cell multi-omics, spatial transcriptomics and systematic perturbation decode circuitry of neural crest fate decisions
Hu Z. et al.
Cranial neural crest (NC) cells, which can migrate, adopt multiple fates, and form most of the craniofacial skeleton, are an excellent model for studying cell fate decisions. Using time-resolved single-cell multi-omics, spatial transcriptomics, and systematic Perturb-seq, we fully deciphered zebrafish cranial NC pro... |
KRAS promotes GLI2-dependent transcription during pancreatic carcinogenesis
Sigafoos A.N. et al.
Aberrant activation of GLI transcription factors has been implicated in the pathogenesis of different tumor types including pancreatic ductal adenocarcinoma (PDAC). However, the mechanistic link with established drivers of this disease remains in part elusive. Here, using a new genetically-engineered mouse model ove... |
Chromatin profiling reveals TFAP4 as a critical transcriptional regulator of bovine satellite cell differentiation
Pengcheng Lyu et al.
Background
Satellite cells are myogenic precursor cells in adult skeletal muscle and play a crucial role in skeletal muscle regeneration, maintenance, and growth. Like embryonic myoblasts, satellite cells have the ability to proliferate, differentiate, and fuse to form multinucleated myofibers. In this study, we ai... |
RUNX1 colludes with NOTCH1 to reprogram chromatin in T-cell acutelymphoblastic leukemia
Islam R. et al.
Runt-related transcription factor 1 (RUNX1) is oncogenic in diverse types of leukemia and epithelial cancers where its expression is associated with poor prognosis. Current models suggest that RUNX1 cooperates with other oncogenic factors (e.g., NOTCH1, TAL1) to drive the expression of proto-oncogenes in T cell... |
Incomplete transcriptional dosage compensation of vertebrate sexchromosomes is balanced by post-transcriptional compensation
Lister N. C. et al.
Heteromorphic sex chromosomes (XY or ZW) present problems of gene dosage imbalance between the sexes, and with the autosomes. Mammalian X chromosome inactivation was long thought to imply a critical need for dosage compensation in vertebrates. However, the universal importance of sex chromosome dosage compensation w... |
Gene Regulatory Interactions at Lamina-Associated Domains
Madsen-Østerbye J. et al.
The nuclear lamina provides a repressive chromatin environment at the nuclear periphery. However, whereas most genes in lamina-associated domains (LADs) are inactive, over ten percent reside in local euchromatic contexts and are expressed. How these genes are regulated and whether they are able to interact with regu... |
Repression and 3D-restructuring resolves regulatory conflicts inevolutionarily rearranged genomes.
Ringel A. et al.
Regulatory landscapes drive complex developmental gene expression, but it remains unclear how their integrity is maintained when incorporating novel genes and functions during evolution. Here, we investigated how a placental mammal-specific gene, Zfp42, emerged in an ancient vertebrate topologically associated domai... |
HOTAIR interacts with PRC2 complex regulating the regional preadipocytetranscriptome and human fat distribution.
Kuo Feng-Chih et al.
Mechanisms governing regional human adipose tissue (AT) development remain undefined. Here, we show that the long non-coding RNA HOTAIR (HOX transcript antisense RNA) is exclusively expressed in gluteofemoral AT, where it is essential for adipocyte development. We find that HOTAIR interacts with polycomb repressive ... |
Local euchromatin enrichment in lamina-associated domains anticipatestheir repositioning in the adipogenic lineage.
Madsen-Østerbye J. et al.
BACKGROUND: Interactions of chromatin with the nuclear lamina via lamina-associated domains (LADs) confer structural stability to the genome. The dynamics of positioning of LADs during differentiation, and how LADs impinge on developmental gene expression, remains, however, elusive. RESULTS: We examined changes in t... |
Loss of KMT2C reprograms the epigenomic landscape in hPSCsresulting in NODAL overexpression and a failure of hemogenic endotheliumspecification.
Maurya Shailendra et al.
Germline or somatic variation in the family of KMT2 lysine methyltransferases have been associated with a variety of congenital disorders and cancers. Notably, -fusions are prevalent in 70\% of infant leukaemias but fail to phenocopy short latency leukaemogenesis in mammalian models, suggesting additional factors ar... |
Functional annotations of three domestic animal genomes provide vitalresources for comparative and agricultural research.
Kern C. et al.
Gene regulatory elements are central drivers of phenotypic variation and thus of critical importance towards understanding the genetics of complex traits. The Functional Annotation of Animal Genomes consortium was formed to collaboratively annotate the functional elements in animal genomes, starting with domesticate... |
Long intergenic non-coding RNAs regulate human lung fibroblast function: Implications for idiopathic pulmonary fibrosis.
Hadjicharalambous MR, Roux BT, Csomor E, Feghali-Bostwick CA, Murray LA, Clarke DL, Lindsay MA
Phenotypic changes in lung fibroblasts are believed to contribute to the development of Idiopathic Pulmonary Fibrosis (IPF), a progressive and fatal lung disease. Long intergenic non-coding RNAs (lincRNAs) have been identified as novel regulators of gene expression and protein activity. In non-stimulated cells, we o... |
Global distribution of DNA hydroxymethylation and DNA methylation in chronic lymphocytic leukemia.
Wernig-Zorc S, Yadav MP, Kopparapu PK, Bemark M, Kristjansdottir HL, Andersson PO, Kanduri C, Kanduri M
BACKGROUND: Chronic lymphocytic leukemia (CLL) has been a good model system to understand the functional role of 5-methylcytosine (5-mC) in cancer progression. More recently, an oxidized form of 5-mC, 5-hydroxymethylcytosine (5-hmC) has gained lot of attention as a regulatory epigenetic modification with prognostic ... |
The Itaconate Pathway Is a Central Regulatory Node Linking Innate Immune Tolerance and Trained Immunity
Domínguez-Andrés Jorge, Novakovic Boris, Li Yang, Scicluna Brendon P., Gresnigt Mark S., Arts Rob J.W., Oosting Marije, Moorlag Simone J.C.F.M., Groh Laszlo A., Zwaag Jelle, Koch Rebecca M., ter Horst Rob, Joosten Leo A.B., Wijmenga Cisca, Michelucci Ales
Sepsis involves simultaneous hyperactivation of the immune system and immune paralysis, leading to both organ dysfunction and increased susceptibility to secondary infections. Acute activation of myeloid cells induced itaconate synthesis, which subsequently mediated innate immune tolerance in human monocytes. In con... |
Increased H3K9 methylation and impaired expression of Protocadherins are associated with the cognitive dysfunctions of the Kleefstra syndrome.
Iacono G, Dubos A, Méziane H, Benevento M, Habibi E, Mandoli A, Riet F, Selloum M, Feil R, Zhou H, Kleefstra T, Kasri NN, van Bokhoven H, Herault Y, Stunnenberg HG
Kleefstra syndrome, a disease with intellectual disability, autism spectrum disorders and other developmental defects is caused in humans by haploinsufficiency of EHMT1. Although EHMT1 and its paralog EHMT2 were shown to be histone methyltransferases responsible for deposition of the di-methylated H3K9 (H3K9me2), th... |
A long range distal enhancer controls temporal fine-tuning of PAX6 expression in neuronal precursors.
Lacomme M, Medevielle F, Bourbon HM, Thierion E, Kleinjan DJ, Roussat M, Pituello F, Bel-Vialar S
Proper embryonic development relies on a tight control of spatial and temporal gene expression profiles in a highly regulated manner. One good example is the ON/OFF switching of the transcription factor PAX6 that governs important steps of neurogenesis. In the neural tube PAX6 expression is initiated in neural proge... |
Inhibition of Methyltransferase Setd7 Allows the In Vitro Expansion of Myogenic Stem Cells with Improved Therapeutic Potential.
Judson RN, Quarta M, Oudhoff MJ, Soliman H, Yi L, Chang CK, Loi G, Vander Werff R, Cait A, Hamer M, Blonigan J, Paine P, Doan LTN, Groppa E, He W, Su L, Zhang RH, Xu P, Eisner C, Low M, Barta I, Lewis CB, Zaph C, Karimi MM, Rando TA, Rossi FM
The development of cell therapy for repairing damaged or diseased skeletal muscle has been hindered by the inability to significantly expand immature, transplantable myogenic stem cells (MuSCs) in culture. To overcome this limitation, a deeper understanding of the mechanisms regulating the transition between ac... |
Metabolic Induction of Trained Immunity through the Mevalonate Pathway.
Bekkering S, Arts RJW, Novakovic B, Kourtzelis I, van der Heijden CDCC, Li Y, Popa CD, Ter Horst R, van Tuijl J, Netea-Maier RT, van de Veerdonk FL, Chavakis T, Joosten LAB, van der Meer JWM, Stunnenberg H, Riksen NP, Netea MG
Innate immune cells can develop long-term memory after stimulation by microbial products during infections or vaccinations. Here, we report that metabolic signals can induce trained immunity. Pharmacological and genetic experiments reveal that activation of the cholesterol synthesis pathway, but not the synthesis of... |
Krox20 hindbrain regulation incorporates multiple modes of cooperation between cis-acting elements
Thierion E. et al.
Developmental genes can harbour multiple transcriptional enhancers that act simultaneously or in succession to achieve robust and precise spatiotemporal expression. However, the mechanisms underlying cooperation between cis-acting elements are poorly documented, notably in vertebrates. The mouse gene Krox20 encodes ... |
A transcription factor pulse can prime chromatin for heritable transcriptional memory
Iberg-Badeaux A. et al.
Short-term and long-term transcriptional memory is the phenomenon whereby the kinetics or magnitude of gene induction is enhanced following a prior induction period. Short-term memory persists within one cell generation or in post-mitotic cells, while long-term memory can survive multiple rounds of cell division. We... |
β-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance
Novakovic B. et al.
Innate immune memory is the phenomenon whereby innate immune cells such as monocytes or macrophages undergo functional reprogramming after exposure to microbial components such as lipopolysaccharide (LPS). We apply an integrated epigenomic approach to characterize the molecular events involved in LPS-induced to... |
Enhancer decommissioning by Snail1-induced competitive displacement of TCF7L2 and down-regulation of transcriptional activators results in EPHB2 silencing
Schnappauf O et al.
Transcriptional silencing is a major cause for the inactivation of tumor suppressor genes, however, the underlying mechanisms are only poorly understood. The EPHB2 gene encodes a receptor tyrosine kinase that controls epithelial cell migration and allocation in intestinal crypts. Through its ability to restrict cell... |
cChIP-seq: a robust small-scale method for investigation of histone modifications
Valensisi C et al.
Background
ChIP-seq is highly utilized for mapping histone modifications that are informative about gene regulation and genome annotations. For example, applying ChIP-seq to histone modifications such as H3K4me1 has facilitated generating epigenomic maps of putative enhancers. This powerful technology, however, i... |
C/EBPα Activates Pre-existing and De Novo Macrophage Enhancers during Induced Pre-B Cell Transdifferentiation and Myelopoiesis
van Oevelen C, Collombet S, Vicent G, Hoogenkamp M, Lepoivre C, Badeaux A, Bussmann L, Sardina JL, Thieffry D, Beato M, Shi Y, Bonifer C, Graf T
Highlights
C/EBPα activates two classes of prospective myeloid enhancers in B cells
Pre-existing enhancers are bound by PU.1 and become hyper-activated by C/EBPα
C/EBPα acts as a pioneer factor with delayed kinetics on de novo enhancers
The two types of enhancers direct myeloid cell fat... |
SNAIL1 combines competitive displacement of ASCL2 and epigenetic mechanisms to rapidly silence the EPHB3 tumor suppressor in colorectal cancer.
Rönsch K, Jägle S, Rose K, Seidl M, Baumgartner F, Freihen V, Yousaf A, Metzger E, Lassmann S, Schüle R, Zeiser R, Michoel T, Hecht A
EPHB3 is a critical cellular guidance factor in the intestinal epithelium and an important tumor suppressor in colorectal cancer (CRC) whose expression is frequently lost at the adenoma-carcinoma transition when tumor cells become invasive. The molecular mechanisms underlying EPHB3 silencing are incompletely underst... |
Interrogation of allelic chromatin states in human cells by high-density ChIP-genotyping.
Light N, Adoue V, Ge B, Chen SH, Kwan T, Pastinen T
Allele-specific (AS) assessment of chromatin has the potential to elucidate specific cis-regulatory mechanisms, which are predicted to underlie the majority of the known genetic associations to complex disease. However, development of chromatin landscapes at allelic resolution has been challenging since sites of var... |
Nuclear ARRB1 induces pseudohypoxia and cellular metabolism reprogramming in prostate cancer
Zecchini V, Madhu B, Russell R, Pértega-Gomes N, Warren A, Gaude E, Borlido J, Stark R, Ireland-Zecchini H, Rao R, Scott H, Boren J, Massie C, Asim M, Brindle K, Griffiths J, Frezza C, Neal DE, Mills IG
Tumour cells sustain their high proliferation rate through metabolic reprogramming, whereby cellular metabolism shifts from oxidative phosphorylation to aerobic glycolysis, even under normal oxygen levels. Hypoxia-inducible factor 1A (HIF1A) is a major regulator of this process, but its activation under normoxic con... |
A novel microscopy-based high-throughput screening method to identify proteins that regulate global histone modification levels.
Baas R, Lelieveld D, van Teeffelen H, Lijnzaad P, Castelijns B, van Schaik FM, Vermeulen M, Egan DA, Timmers HT, de Graaf P
Posttranslational modifications of histones play an important role in the regulation of gene expression and chromatin structure in eukaryotes. The balance between chromatin factors depositing (writers) and removing (erasers) histone marks regulates the steady-state levels of chromatin modifications. Here we describe... |
An In-Depth Characterization of the Major Psoriasis Susceptibility Locus Identifies Candidate Susceptibility Alleles within an HLA-C Enhancer Element.
Clop A, Bertoni A, Spain SL, Simpson MA, Pullabhatla V, Tonda R, Hundhausen C, Di Meglio P, De Jong P, Hayday AC, Nestle FO, Barker JN, Bell RJ, Capon F, Trembath RC
Psoriasis is an immune-mediated skin disorder that is inherited as a complex genetic trait. Although genome-wide association scans (GWAS) have identified 36 disease susceptibility regions, more than 50% of the genetic variance can be attributed to a single Major Histocompatibility Complex (MHC) locus, known as PSORS... |
Disease-Related Growth Factor and Embryonic Signaling Pathways Modulate an Enhancer of TCF21 Expression at the 6q23.2 Coronary Heart Disease Locus.
Miller CL, Anderson DR, Kundu RK, Raiesdana A, Nürnberg ST, Diaz R, Cheng K, Leeper NJ, Chen CH, Chang IS, Schadt EE, Hsiung CA, Assimes TL, Quertermous T
Coronary heart disease (CHD) is the leading cause of mortality in both developed and developing countries worldwide. Genome-wide association studies (GWAS) have now identified 46 independent susceptibility loci for CHD, however, the biological and disease-relevant mechanisms for these associations remain elusive. Th... |
Balancing of histone H3K4 methylation states by the Kdm5c/SMCX histone demethylase modulates promoter and enhancer function.
Outchkourov NS, Muiño JM, Kaufmann K, van Ijcken WF, Groot Koerkamp MJ, van Leenen D, de Graaf P, Holstege FC, Grosveld FG, Timmers HT
The functional organization of eukaryotic genomes correlates with specific patterns of histone methylations. Regulatory regions in genomes such as enhancers and promoters differ in their extent of methylation of histone H3 at lysine-4 (H3K4), but it is largely unknown how the different methylation states are specifi... |
Characterization of the contradictory chromatin signatures at the 3' exons of zinc finger genes.
Blahnik KR, Dou L, Echipare L, Iyengar S, O'Geen H, Sanchez E, Zhao Y, Marra MA, Hirst M, Costello JF, Korf I, Farnham PJ
The H3K9me3 histone modification is often found at promoter regions, where it functions to repress transcription. However, we have previously shown that 3' exons of zinc finger genes (ZNFs) are marked by high levels of H3K9me3. We have now further investigated this unusual location for H3K9me3 in ZNF genes. Neither ... |
Using ChIP-Seq Technology to Generate High-Resolution Profiles of Histone Modifications
O’Geen H, Echipare L, Farnham PJ
The dynamic modification of DNA and histones plays a key role in transcriptional regulation through - altering the packaging of DNA and modifying the nucleosome surface. These chromatin states, also referred to as the epigenome, are distinctive for different tissues, developmental stages, and disease states and can ... |