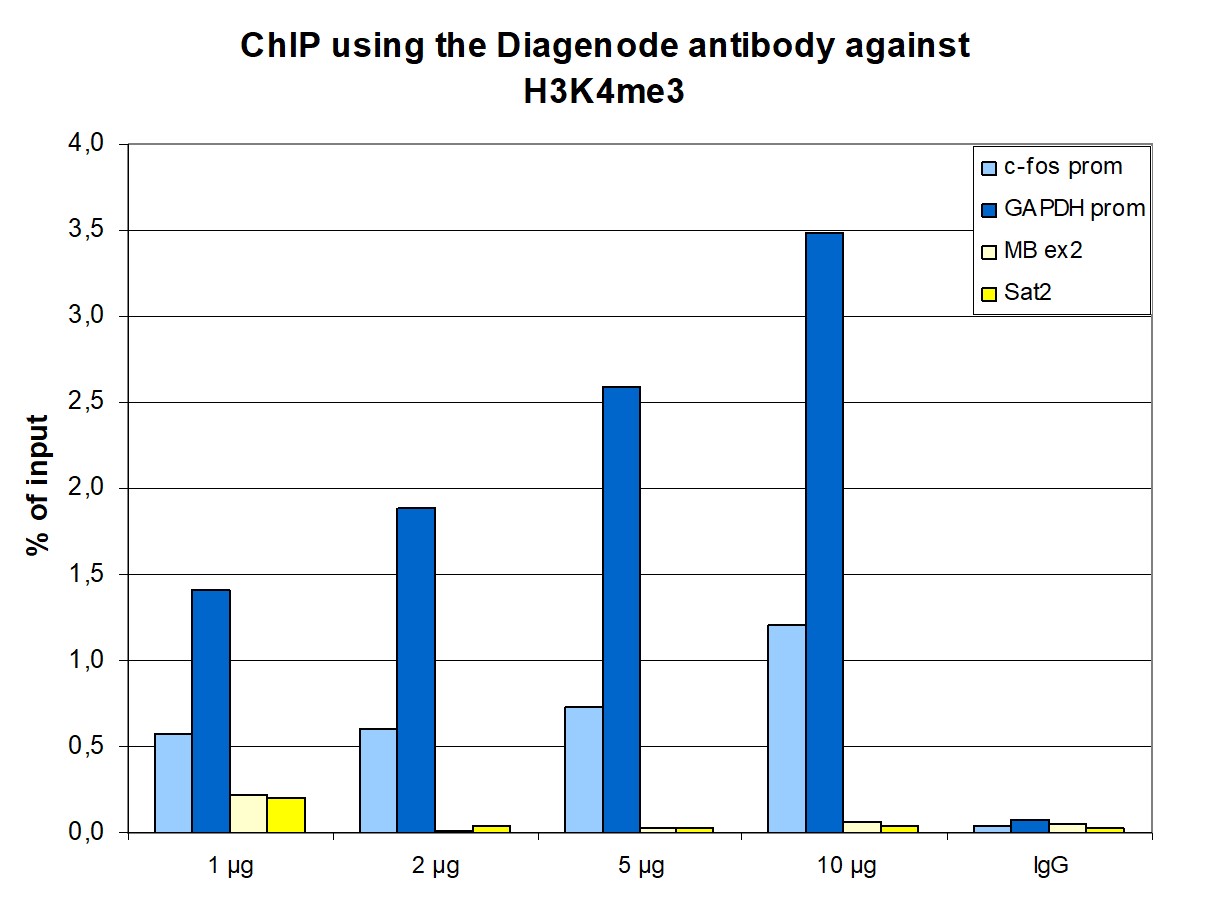
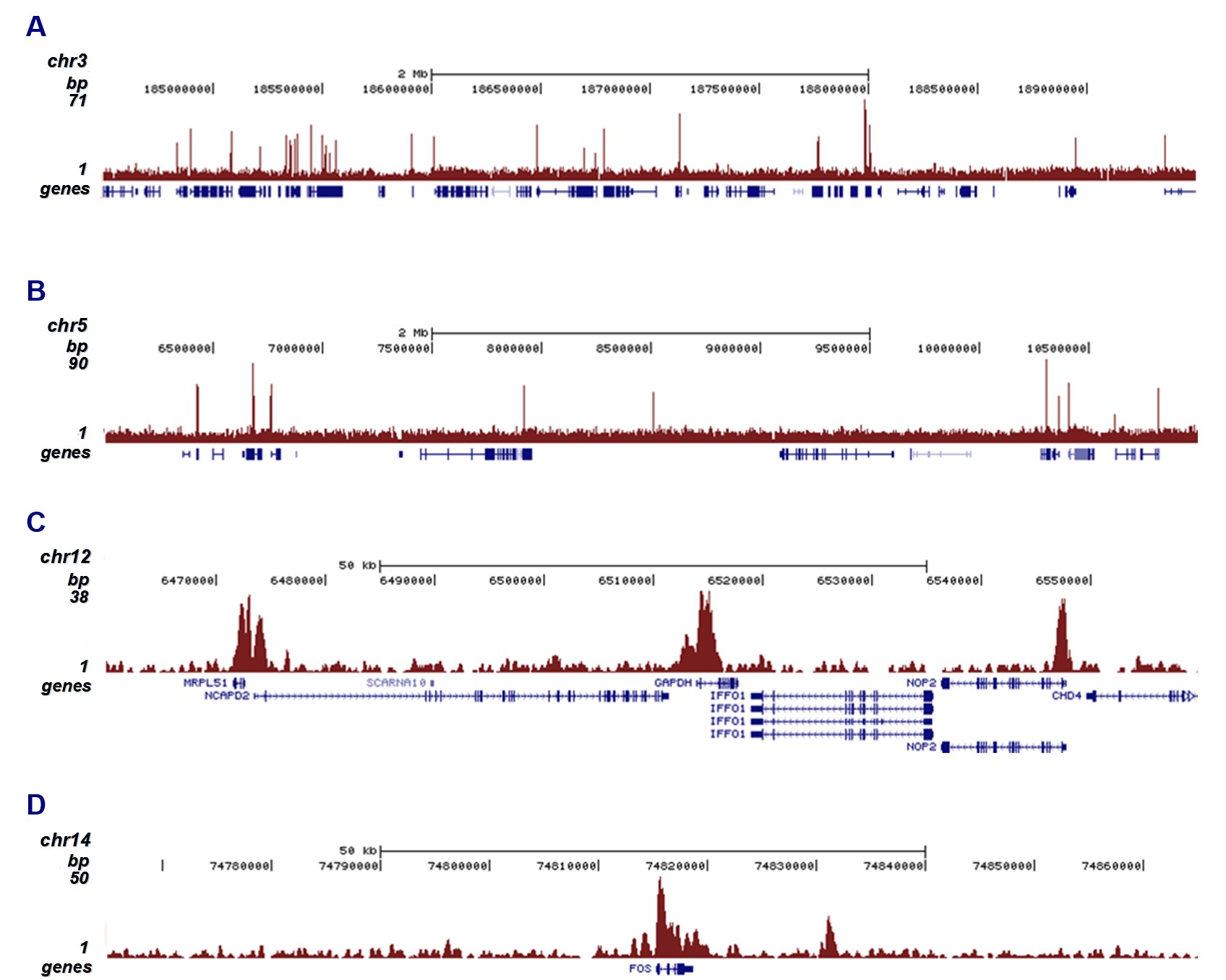
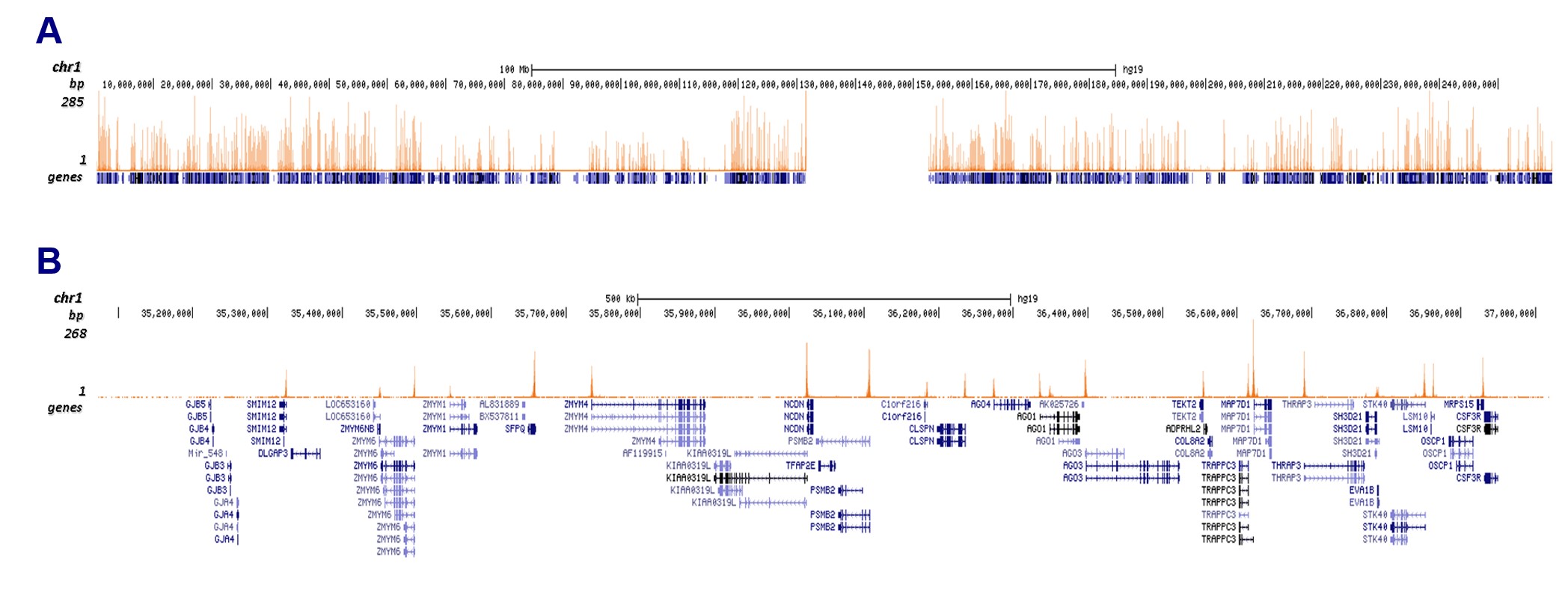
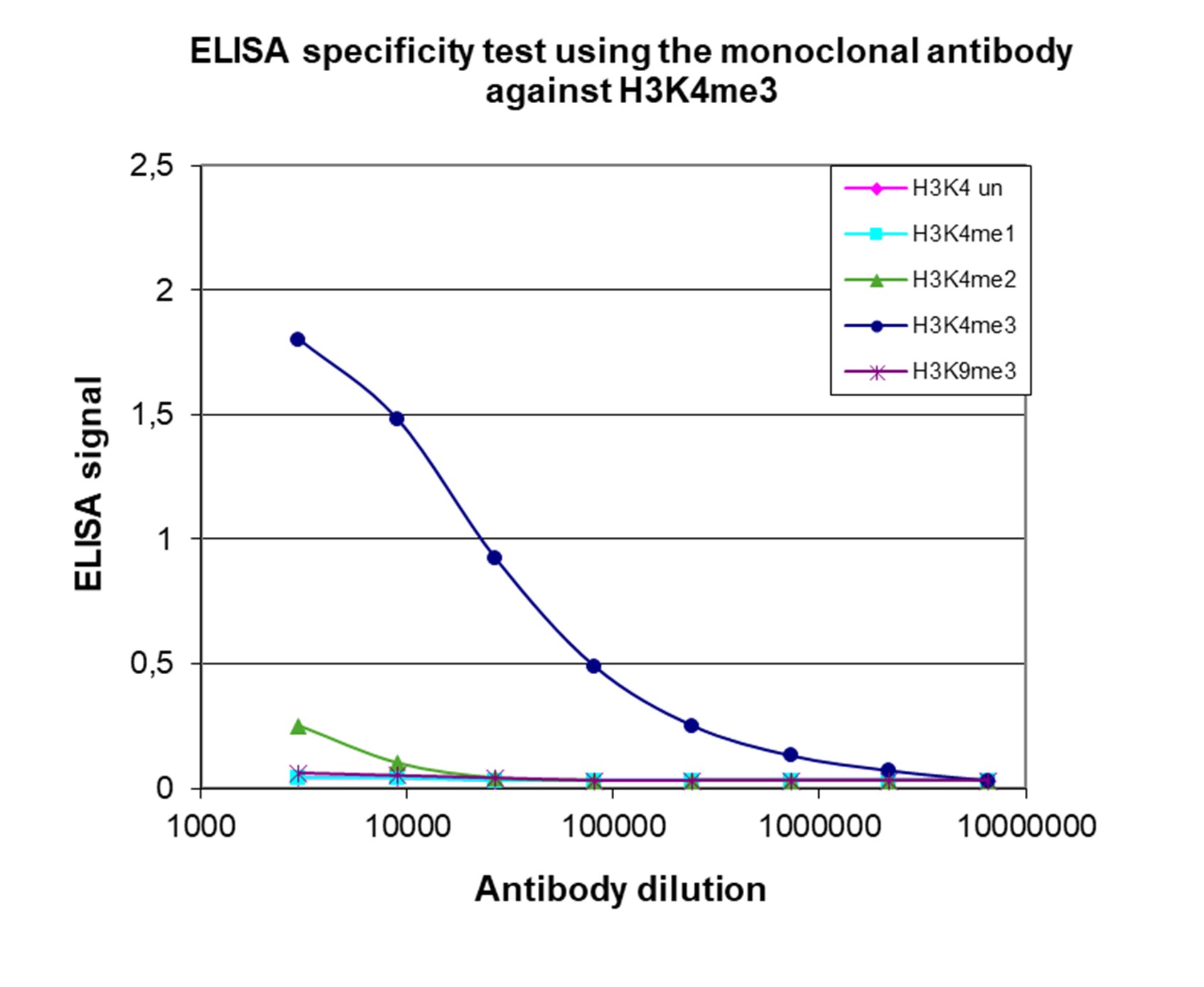
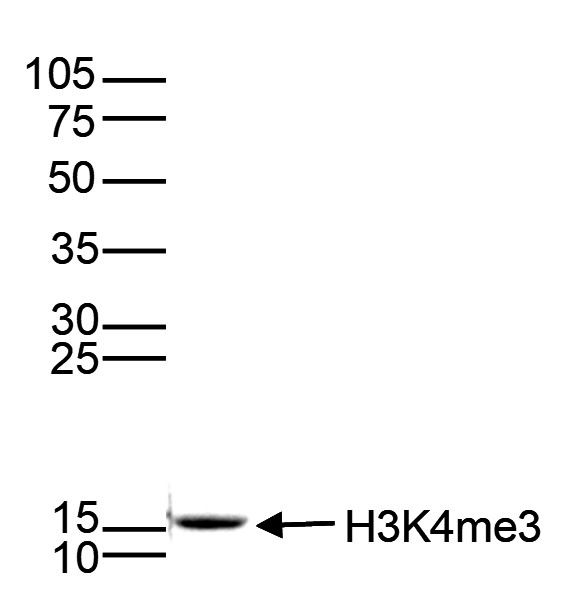
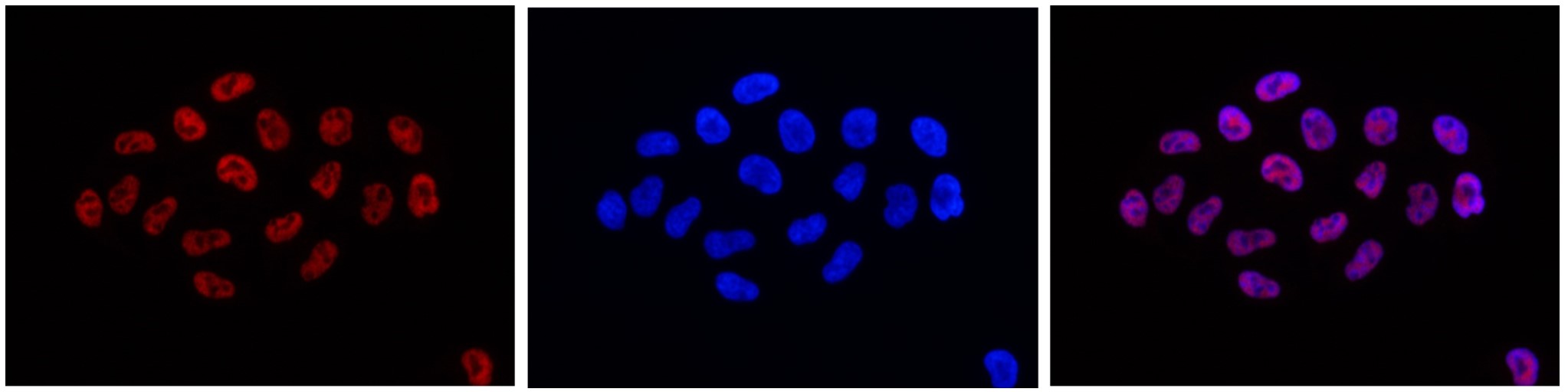
Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3, and H4 assemble and are wrapped by 146 base pairs of DNA to form an octameric nucleosome. Histone tails undergo numerous posttranslational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code.” Histone methylation and demethylation are dynamically regulated by histone methyltransferases and histone demethylases, respectively.