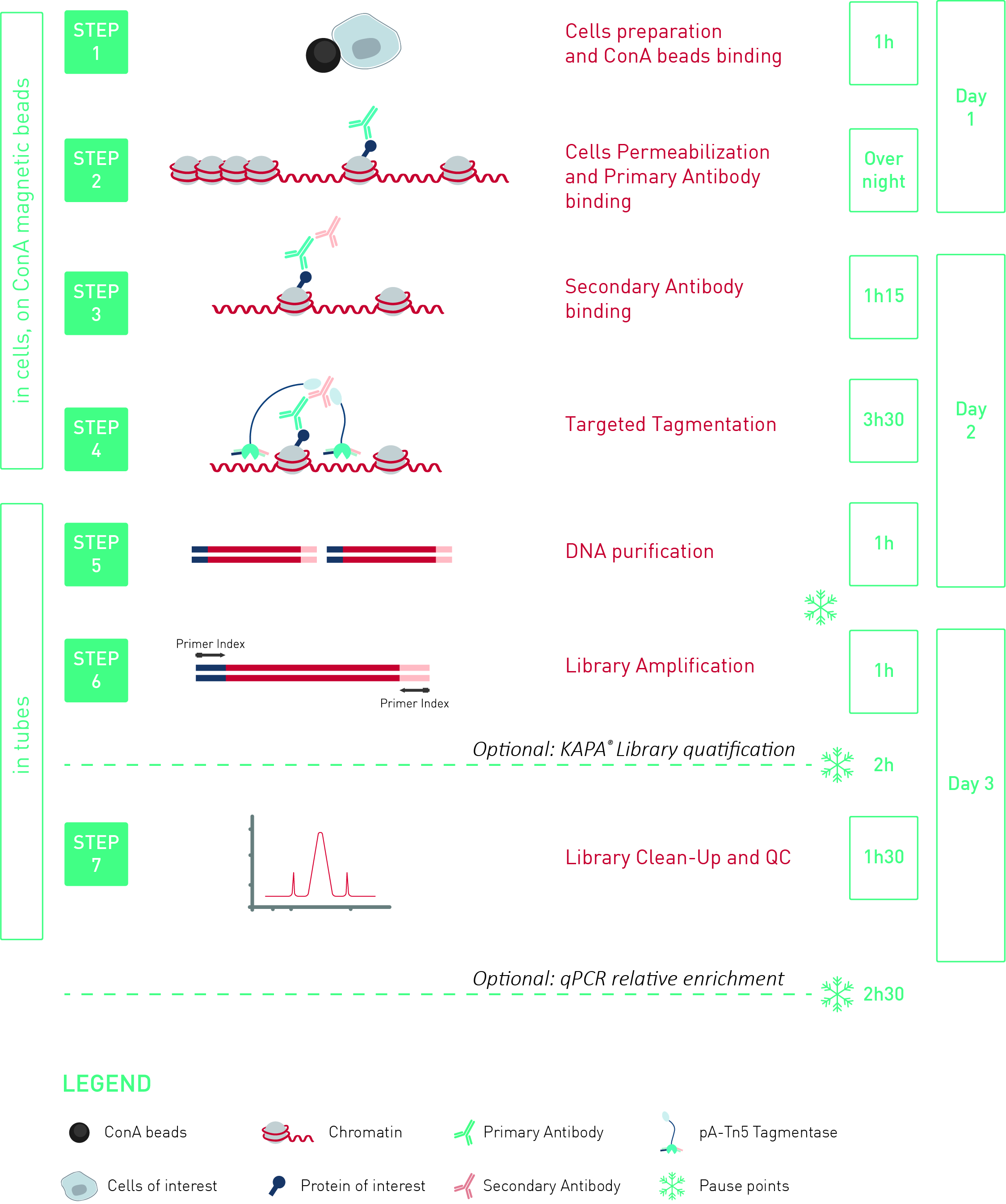
Diagenode's iDeal CUT&Tag Kit for Histones, developed for an efficient chromatin profiling on histone marks, can be used for profiling of some transcription factors and co-factors using a mild fixation as described in the manual.
Successful CUT&Tag results using CTCF and Suz12 antibodies and iDeal CUT&Tag Kit for Histones.
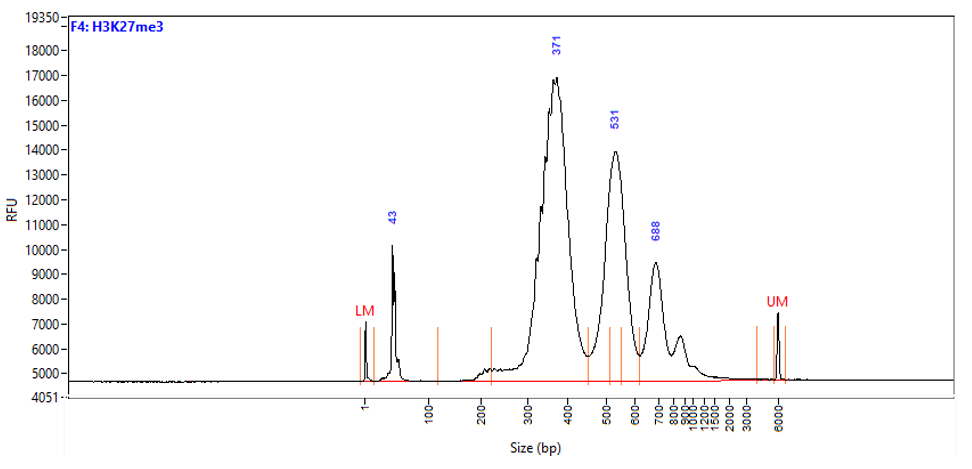
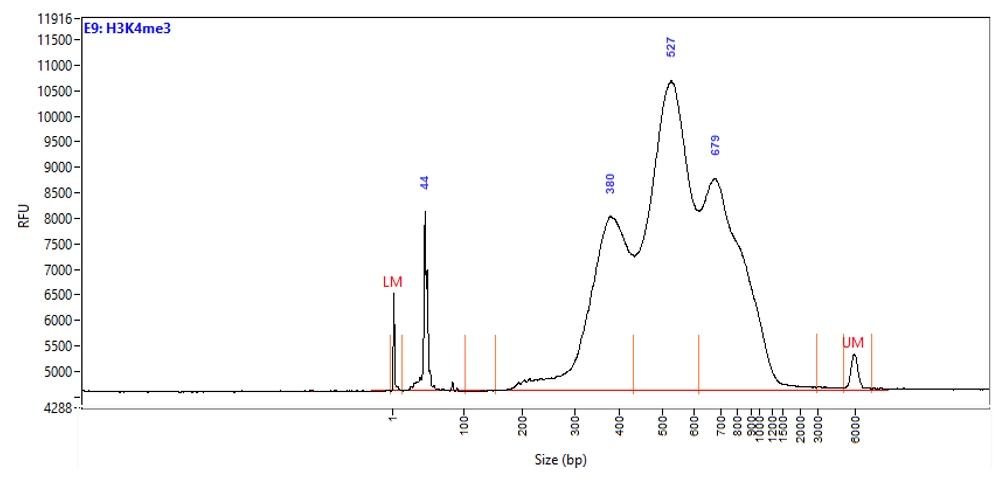
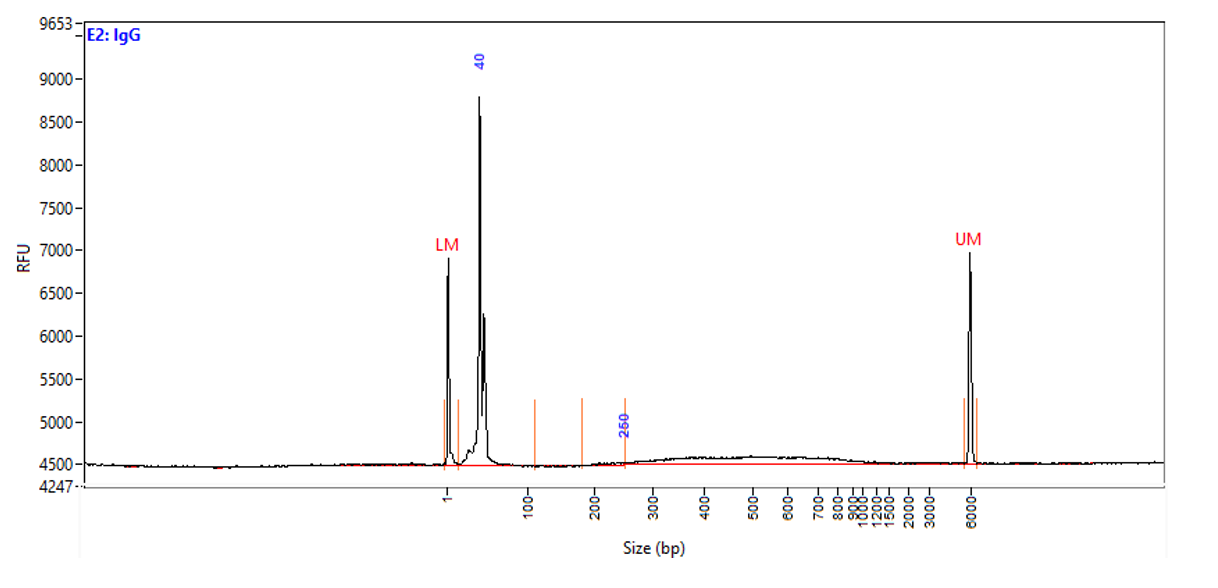
Chromatin profiling of Suz12 has been performed on 50,000 of fixed Mouse ES-E14TG2a cells (kindly provided by Luciano Di Croce, CRG, Spain) accordingly to the protocol of Diagenode’s iDeal CUT&Tag kit for Histones (Cat. No. C01070020). Antibody Package for CUT&Tag anti rabbit (Cat. No. C01070022) and the 24 UDI for Tagmented Libraries (Cat. No. C01011034) were used. The libraries were sized using Fragment Analyzer (Agilent) (triplicates, Figure 1, top). Relative enrichment has been confirmed by qPCR using know positive (T_Bra) and negative (Actb) loci (Figure 1, bottom). Libraries were sequenced on Illumina’s NovaSeq6000 in 2x50 bp mode.
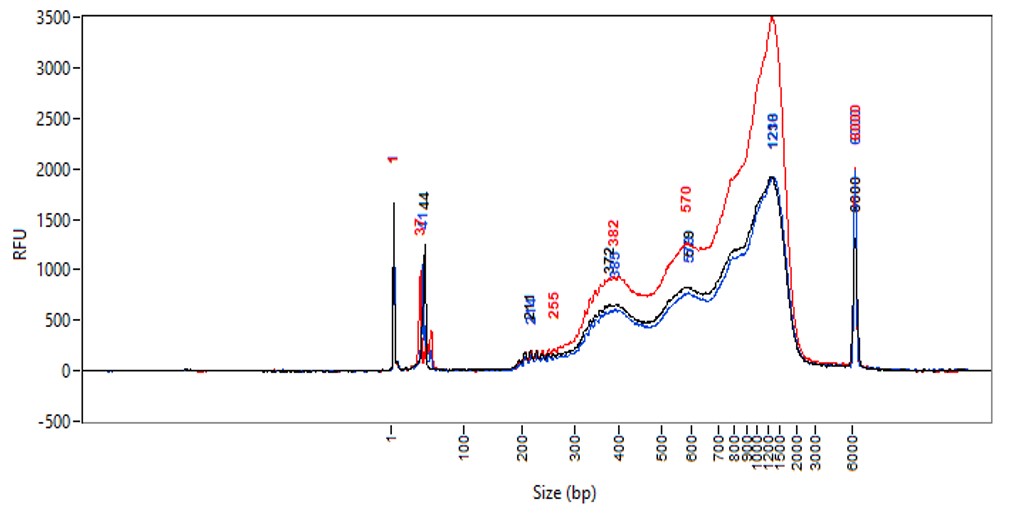
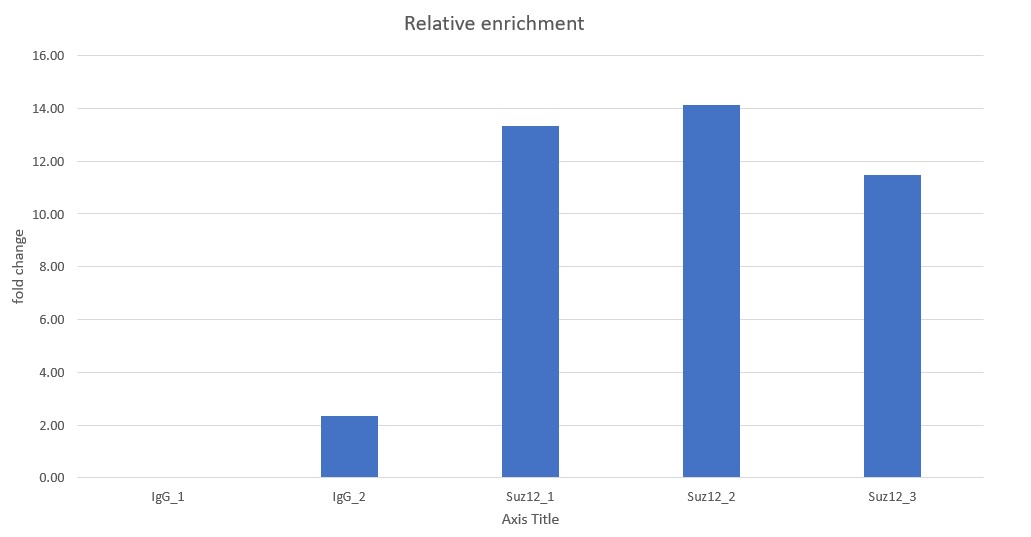
Figure 1. Typical library profiles (top) and relative enrichment (bottom) generated by the iDeal CUT&Tag protocol using 50,000 ES-E14TG2a cells and Suz12 Antibody and IgG control (bottom). Sharp peak at around 40 bp is an excess of free oligonucleotide used for pA-Tn5 loading which does not interfere with the sequencing.
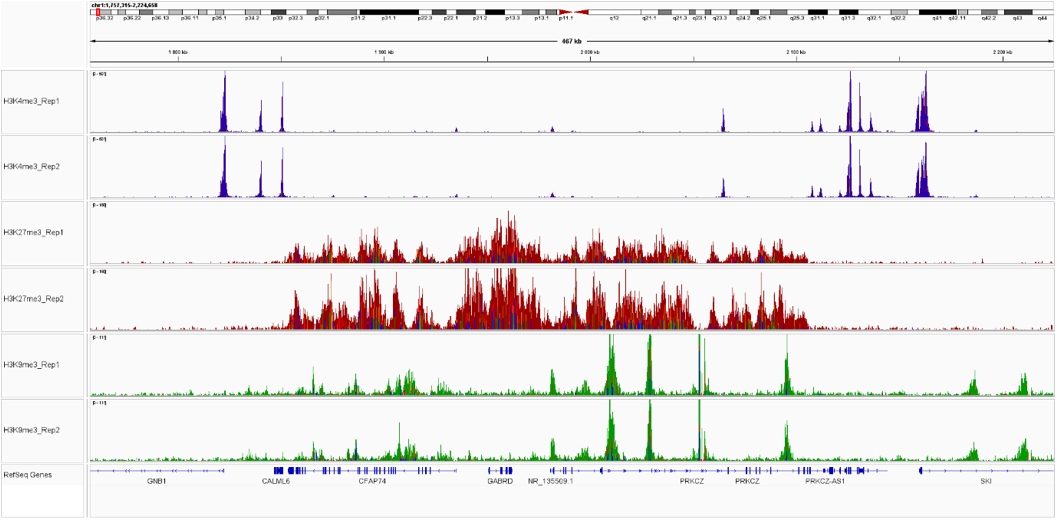
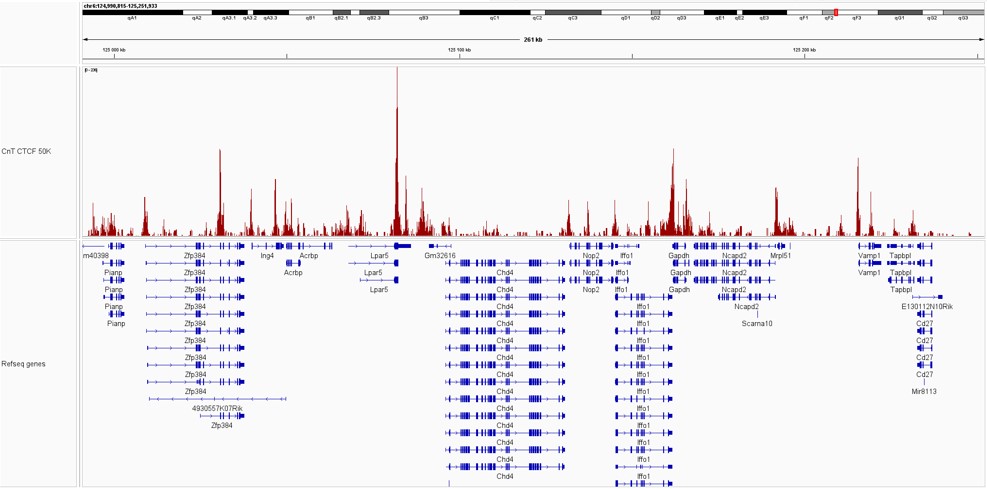
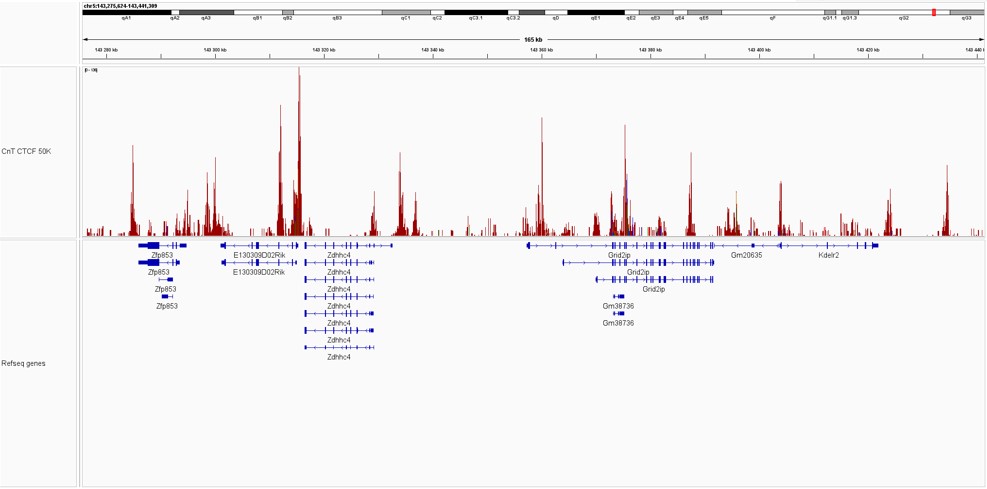
Pictures of representative loci were taken on Integrative Genome Viewer (IGV) for CTCF demonstrating the presence of high signal at promoter regions.
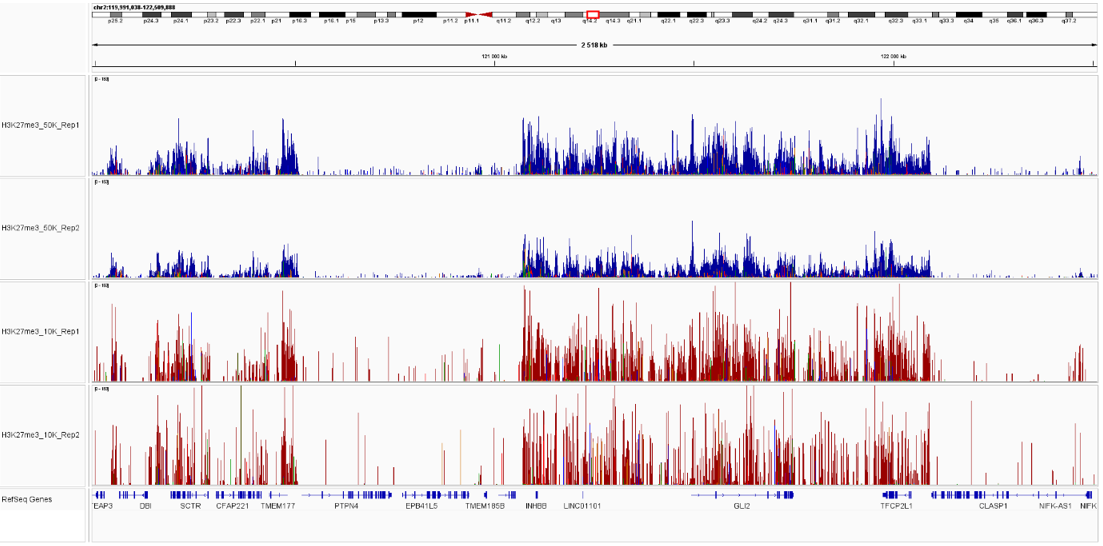
Pictures of representative loci were taken on Integrative Genome Viewer (IGV) for SUZ12 and H3K27me3 triplicate samples, showing an overlap between the signal from both proteins, as part of the Polycomb Repressive Complex.
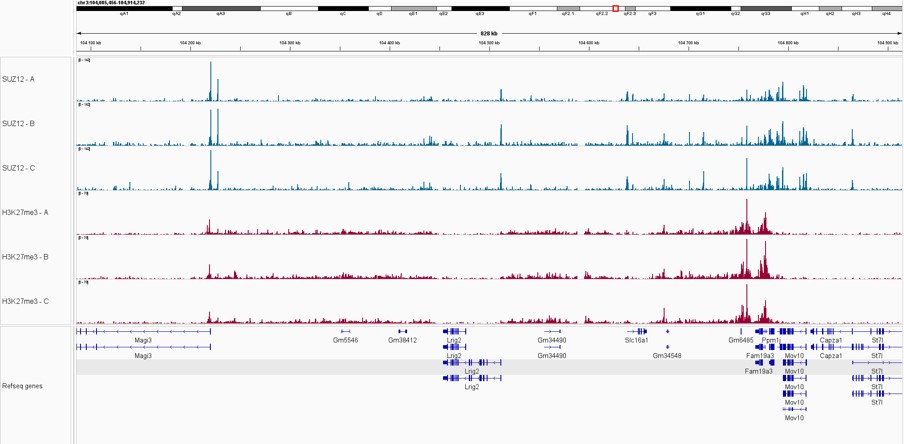
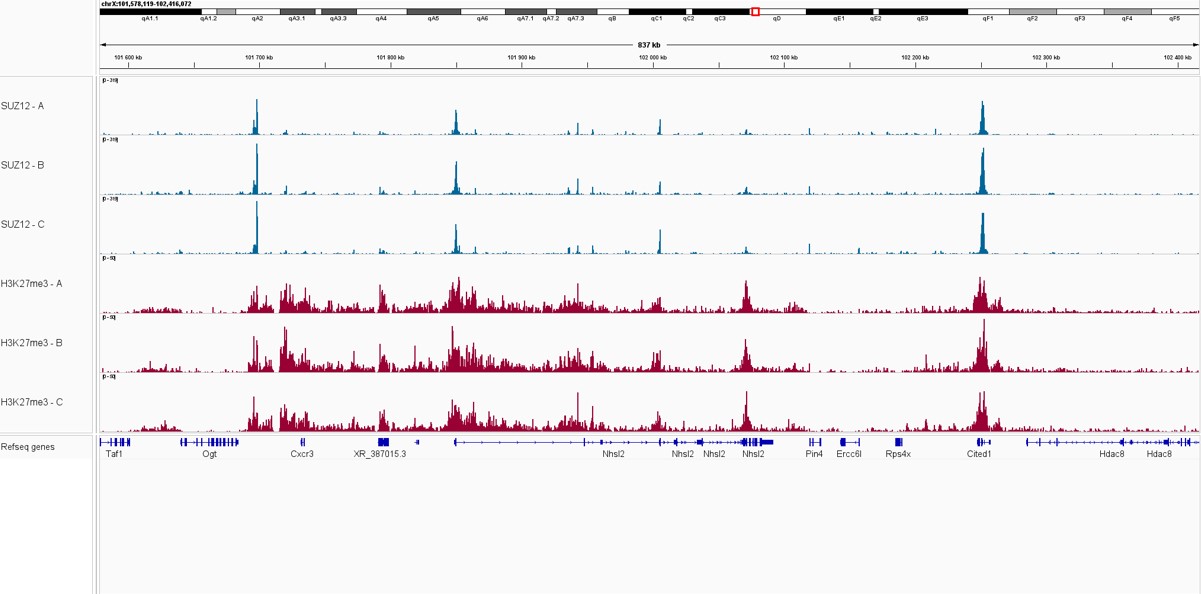
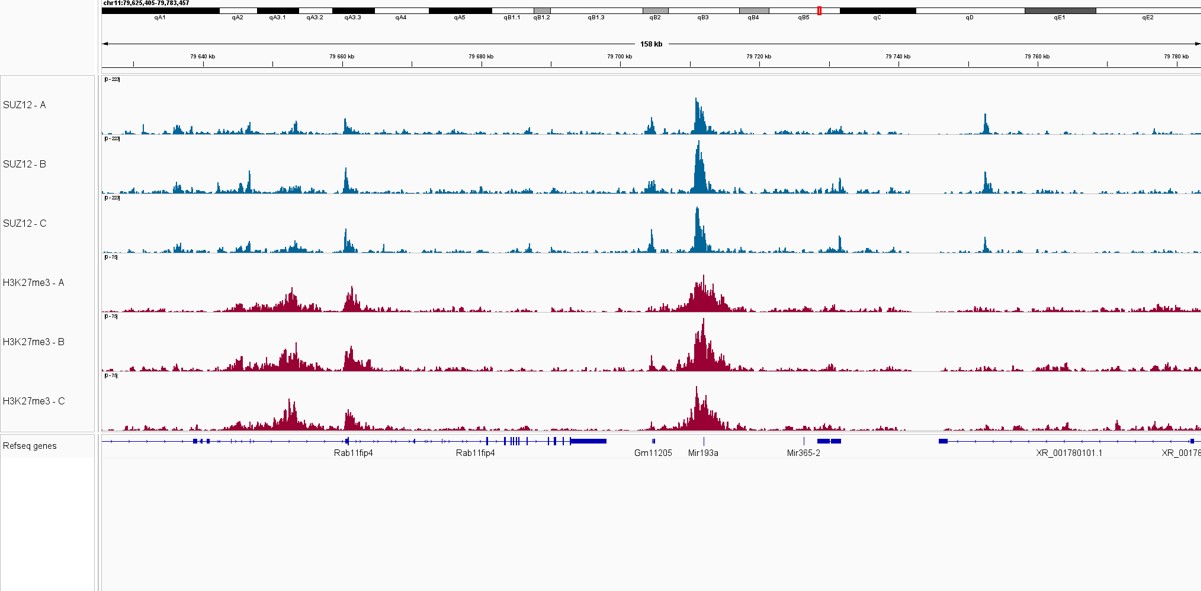
Figure 2. IGV snapshots
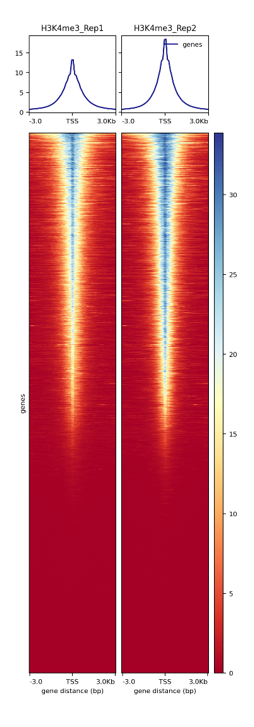
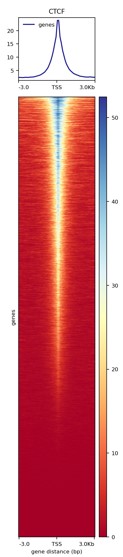
Figure 3. Heatmap around the TSS
Heatmap of the CTCF signal around the transcription start sites (TSS) of each gene present in the murine mm10 reference genome.
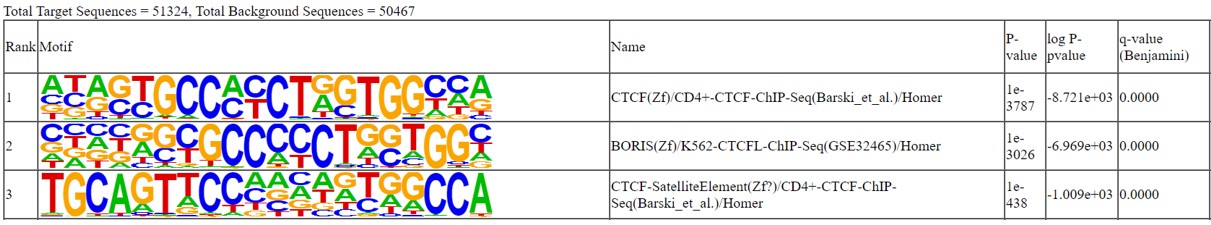
Figure 4. CTCF motif
Presence of specific transcription factor binding motifs in the regions identified as CTCF peaks. The top3 motifs are corresponding to the CTCF binding motif.
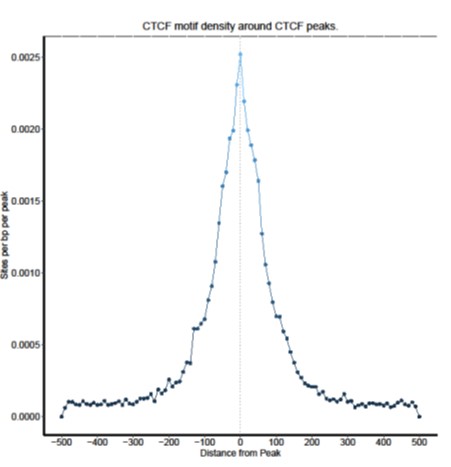
Figure 5. CTCF motif density
Location and density of the CTCF binding motif with respect to the center of the identified CTCF peaks.