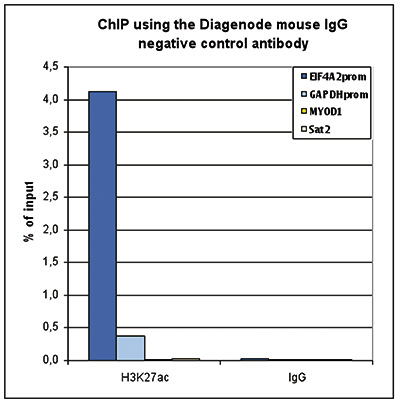
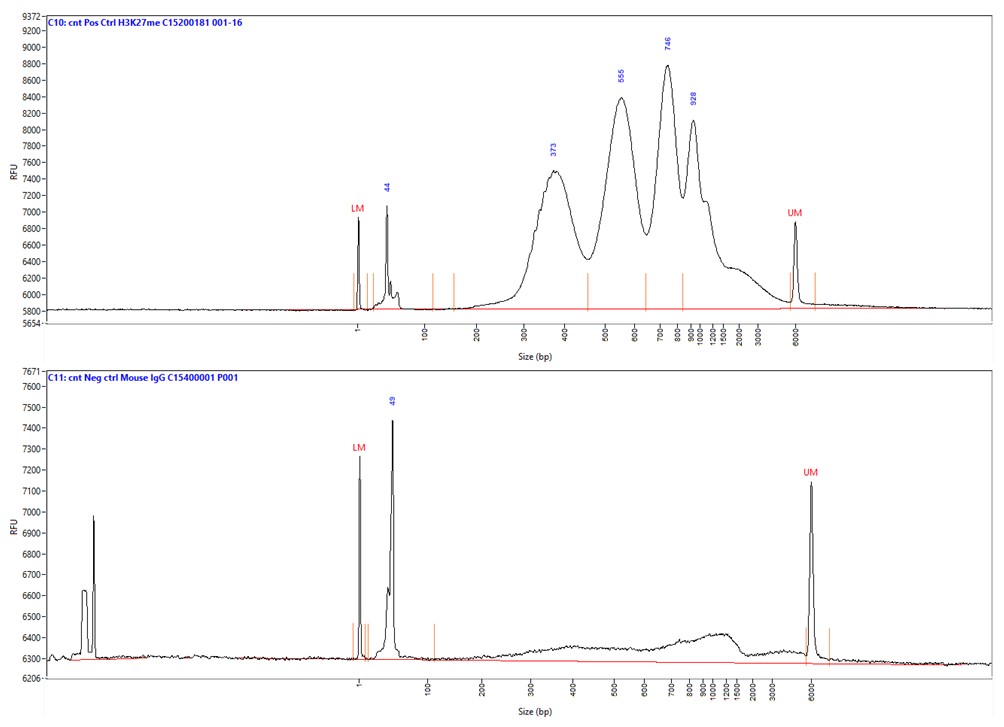
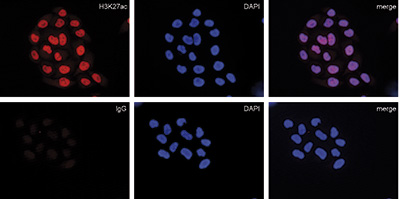
How to properly cite our product/service in your work We strongly recommend using this: Mouse IgG (Hologic Diagenode Cat# C15400001-15 Lot# MIG004C). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Endurance training increases a ubiquitylated form of histone H3 in the skeletal muscle, supporting Notch1 upregulation in an MDM2-dependent manner
Lam, Brian et al.
At the onset of training, each exercise session transiently shifts the distribution of histone post-transcriptional modifications (HPTMs) to activate genes that drive muscle adaptations. The resulting cyclic changes in gene expression promote the acquisition of high oxidative capacities and gains in capillaries. I... |
ETV2/ER71 regulates hematovascular lineage generation and vascularization through an H3K9 demethylase, KDM4A
Min Seong Kim et al.
Highlights
Interaction of ETV2 and KDM4A decreases H3K9 trimethylation on hematovascular genes.
ETV2 and KDM4A cooperatively regulates the expression of hematovascular genes.
Mice lacking endothelial Etv2 and Kdm4a display a severe angiogenic impairment.
... |
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2
Goradia N. et al.
While the elucidation of regulatory mechanisms of folded proteins is facilitated due to their amenability to high-resolution structural characterization, investigation of these mechanisms in disordered proteins is more challenging due to their structural heterogeneity, which can be captured by a variety of biophysic... |
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2
Nishit Goradia et al.
While the elucidation of regulatory mechanisms of folded proteins is facilitated due to their amenability to high-resolution structural characterization, investigation of these mechanisms in disordered proteins is more challenging due to their structural heterogeneity, which can be captured by a variety of biophysic... |
The Effect of Metformin and Carbohydrate-Controlled Diet onDNA Methylation and Gene Expression in the Endometrium of Womenwith Polycystic Ovary Syndrome.
Garcia-Gomez E. et al.
Polycystic ovary syndrome (PCOS) is an endocrine disease associated with infertility and metabolic disorders in reproductive-aged women. In this study, we evaluated the expression of eight genes related to endometrial function and their DNA methylation levels in the endometrium of PCOS patients and women without the... |
The histone modification H3K4me3 is altered at the locus in Alzheimer'sdisease brain.
Smith, Adam et al.
Several epigenome-wide association studies of DNA methylation have highlighted altered DNA methylation in the gene in Alzheimer's disease (AD) brain samples. However, no study has specifically examined histone modifications in the disease. We use chromatin immunoprecipitation-qPCR to quantify tri-methylation at hist... |
Inhibition of HIV-1 gene transcription by KAP1 in myeloid lineage.
Ait-Ammar A. et al.
HIV-1 latency generates reservoirs that prevent viral eradication by the current therapies. To find strategies toward an HIV cure, detailed understandings of the molecular mechanisms underlying establishment and persistence of the reservoirs are needed. The cellular transcription factor KAP1 is known as a potent rep... |
p53 directly represses human LINE1 transposons.
Tiwari, Bhavana and Jones, Amanda E and Caillet, Candace J and Das, Simantiand Royer, Stephanie K and Abrams, John M
p53 is a potent tumor suppressor and commonly mutated in human cancers. Recently, we demonstrated that p53 genes act to restrict retrotransposons in germline tissues of flies and fish but whether this activity is conserved in somatic human cells is not known. Here we show that p53 constitutively restrains human LINE... |
Attenuated Epigenetic Suppression of Muscle Stem Cell Necroptosis Is Required for Efficient Regeneration of Dystrophic Muscles.
Sreenivasan K, Ianni A, Künne C, Strilic B, Günther S, Perdiguero E, Krüger M, Spuler S, Offermanns S, Gómez-Del Arco P, Redondo JM, Munoz-Canoves P, Kim J, Braun T
Somatic stem cells expand massively during tissue regeneration, which might require control of cell fitness, allowing elimination of non-competitive, potentially harmful cells. How or if such cells are removed to restore organ function is not fully understood. Here, we show that a substantial fraction of muscle stem... |
Episo: quantitative estimation of RNA 5-methylcytosine at isoform level by high-throughput sequencing of RNA treated with bisulfite.
Liu J, An Z, Luo J, Li J, Li F, Zhang Z
MOTIVATION: RNA 5-methylcytosine (m5C) is a type of post-transcriptional modification that may be involved in numerous biological processes and tumorigenesis. RNA m5C can be profiled at single-nucleotide resolution by high-throughput sequencing of RNA treated with bisulfite (RNA-BisSeq). However, the exploration of ... |
Centromeres License the Mitotic Condensation of Yeast Chromosome Arms.
Kruitwagen T, Chymkowitch P, Denoth-Lippuner A, Enserink J, Barral Y
During mitosis, chromatin condensation shapes chromosomes as separate, rigid, and compact sister chromatids to facilitate their segregation. Here, we show that, unlike wild-type yeast chromosomes, non-chromosomal DNA circles and chromosomes lacking a centromere fail to condense during mitosis. The centromere promote... |
A novel FOXA1/ESR1 interacting pathway: A study of Oncomine™ breast cancer microarrays
Chaudhary S. et al.
Forkhead box protein A1 (FOXA1) is essential for the growth and differentiation of breast epithelium, and has a favorable outcome in breast cancer (BC). Elevated FOXA1 expression in BC also facilitates hormone responsiveness in estrogen receptor (ESR)-positive BC. However, the interaction between these two pathways ... |
TET-Catalyzed 5-Hydroxymethylation Precedes HNF4A Promoter Choice during Differentiation of Bipotent Liver Progenitors
Ancey P.B. et al.
Understanding the processes that govern liver progenitor cell differentiation has important implications for the design of strategies targeting chronic liver diseases, whereby regeneration of liver tissue is critical. Although DNA methylation (5mC) and hydroxymethylation (5hmC) are highly dynamic during early embryo... |
Differentiation of Mouse Enteric Nervous System Progenitor Cells is Controlled by Endothelin 3 and Requires Regulation of Ednrb by SOX10 and ZEB2
Watanabe Y. et al.
Background & Aims
Maintenance and differentiation of progenitor cells in the developing enteric nervous system (ENS) are controlled by molecules such as the signaling protein endothelin 3 (EDN3), its receptor (the endothelin receptor type B, EDNRB), and the transcription factors SRY-box 10 (SOX10) and zinc fi... |
High resolution methylation analysis of the HoxA5 regulatory region in different somatic tissues of laboratory mouse during development
Sinha P. et al.
Homeobox genes encode a group of DNA binding regulatory proteins whose key function occurs in the spatial-temporal organization of genome during embryonic development and differentiation. The role of these Hox genes during ontogenesis makes it an important model for research. HoxA5 is a member of Hox gene family pla... |
Overexpression of histone demethylase Fbxl10 leads to enhanced migration in mouse embryonic fibroblasts.
Rohde M. et al.
Cell migration is a central process in the development and maintenance of multicellular organisms. Tissue formation during embryonic development, wound healing, immune responses and invasive tumors all require the orchestrated movement of cells to specific locations. Histone demethylase proteins alter transcription ... |
Methylation of the Sox9 and Oct4 promoters and its correlation with gene expression during testicular development in the laboratory mouse
Pamnani M et al.
Sox9 and Oct4 are two important regulatory factors involved in mammalian development. Sox9, a member of the group E Sox transcription factor family, has a crucial role in the development of the genitourinary system, while Oct4, commonly known as octamer binding transcription factor 4, belongs to class V of the trans... |
Overexpression of caspase 7 is ERα dependent to affect proliferation and cell growth in breast cancer cells by targeting p21(Cip)
Chaudhary S, Madhukrishna B, Adhya AK, Keshari S, Mishra SK
Caspase 7 (CASP7) expression has important function during cell cycle progression and cell growth in certain cancer cells and is also involved in the development and differentiation of dental tissues. However, the function of CASP7 in breast cancer cells is unclear. The aim of this study was to analyze the expressio... |
Methylated DNA Immunoprecipitation Analysis of Mammalian Endogenous Retroviruses.
Rebollo R, Mager DL
Endogenous retroviruses are repetitive sequences found abundantly in mammalian genomes which are capable of modulating host gene expression. Nevertheless, most endogenous retrovirus copies are under tight epigenetic control via histone-repressive modifications and DNA methylation. Here we describe a common method us... |
Chromatin Immunoprecipitation Assay for the Identification of Arabidopsis Protein-DNA Interactions In Vivo
Komar DN, Mouriz A, Jarillo JA, Piñeiro M
Intricate gene regulatory networks orchestrate biological processes and developmental transitions in plants. Selective transcriptional activation and silencing of genes mediate the response of plants to environmental signals and developmental cues. Therefore, insights into the mechanisms that control plant gene expr... |
The genetic association of RUNX3 with ankylosing spondylitis can be explained by allele-specific effects on IRF4 recruitment that alter gene expression
Matteo Vecellio, Amity R Roberts, Carla J Cohen, Adrian Cortes, Julian C Knight, Paul Bowness, B Paul Wordsworth
The authors sought to identify the functional basis for the genetic association of single nucleotide polymorphisms (SNP), upstream of the RUNX3 promoter, with ankylosing spondylitis (AS). They performed conditional analysis of genetic association data and used ENCODE data on chromatin remodelling and transcription f... |
ClpX stimulates the mitochondrial unfolded protein response (UPRmt) in mammalian cells
Al-Furoukh N, Ianni A, Nolte H, Hölper S, Krüger M, Wanrooij S, Braun T
Proteostasis is crucial for life and maintained by cellular chaperones and proteases. One major mitochondrial protease is the ClpXP complex, which is comprised of a catalytic ClpX subunit and a proteolytic ClpP subunit. Based on two separate observations, we hypothesized that ClpX may play a leading role in the cell... |
Endothelial Msx1 transduces hemodynamic changes into an arteriogenic remodeling response.
Vandersmissen I, Craps S, Depypere M, Coppiello G, van Gastel N, Maes F, Carmeliet G, Schrooten J, Jones EA, Umans L, Devlieger R, Koole M, Gheysens O, Zwijsen A, Aranguren XL, Luttun A
Collateral remodeling is critical for blood flow restoration in peripheral arterial disease and is triggered by increasing fluid shear stress in preexisting collateral arteries. So far, no arterial-specific mediators of this mechanotransduction response have been identified. We show that muscle segment homeobox 1 (M... |
DNA methylation directs functional maturation of pancreatic β cells
Dhawan S, Tschen SI, Zeng C, Guo T, Hebrok M, Matveyenko A, Bhushan A.
Pancreatic β cells secrete insulin in response to postprandial increases in glucose levels to prevent hyperglycemia and inhibit insulin secretion under fasting conditions to protect against hypoglycemia. β cells lack this functional capability at birth and acquire glucose-stimulated insulin secretion (GSIS... |
Trrap-dependent histone acetylation specifically regulates cell-cycle gene transcription to control neural progenitor fate decisions.
Tapias A, Zhou ZW, Shi Y, Chong Z, Wang P, Groth M, Platzer M, Huttner W, Herceg Z, Yang YG, Wang ZQ
Fate decisions in neural progenitor cells are orchestrated via multiple pathways, and the role of histone acetylation in these decisions has been ascribed to a general function promoting gene activation. Here, we show that the histone acetyltransferase (HAT) cofactor transformation/transcription domain-associated pr... |
Alu Elements in ANRIL Non-Coding RNA at Chromosome 9p21 Modulate Atherogenic Cell Functions through Trans-Regulation of Gene Networks.
Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gäbel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D
The chromosome 9p21 (Chr9p21) locus of coronary artery disease has been identified in the first surge of genome-wide association and is the strongest genetic factor of atherosclerosis known today. Chr9p21 encodes the long non-coding RNA (ncRNA) antisense non-coding RNA in the INK4 locus (ANRIL). ANRIL expression is ... |
Genome-wide analysis of LXRα activation reveals new transcriptional networks in human atherosclerotic foam cells.
Feldmann R, Fischer C, Kodelja V, Behrens S, Haas S, Vingron M, Timmermann B, Geikowski A, Sauer S
Increased physiological levels of oxysterols are major risk factors for developing atherosclerosis and cardiovascular disease. Lipid-loaded macrophages, termed foam cells, are important during the early development of atherosclerotic plaques. To pursue the hypothesis that ligand-based modulation of the nuclear recep... |
New partners in regulation of gene expression: the enhancer of trithorax and polycomb corto interacts with methylated ribosomal protein l12 via its chromodomain.
Coléno-Costes A, Jang SM, de Vanssay A, Rougeot J, Bouceba T, Randsholt NB, Gibert JM, Le Crom S, Mouchel-Vielh E, Bloyer S, Peronnet F
Chromodomains are found in many regulators of chromatin structure, and most of them recognize methylated lysines on histones. Here, we investigate the role of the Drosophila melanogaster protein Corto's chromodomain. The Enhancer of Trithorax and Polycomb Corto is involved in both silencing and activation of gene ex... |
The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology and the metabolome of fibroblasts
Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J
Fbxl10 (Jhdm1b/Kdm2b) is a conserved and ubiquitously expressed member of the JHDM (JmjC-domain-containing histone demethy-lase) family. Fbxl10 was implicated in the demethylation of H3K4me3 or H3K36me2 thereby removing active chromatin marks and inhibiting gene transcription. Apart from the JmjC domain, Fbxl10 cons... |