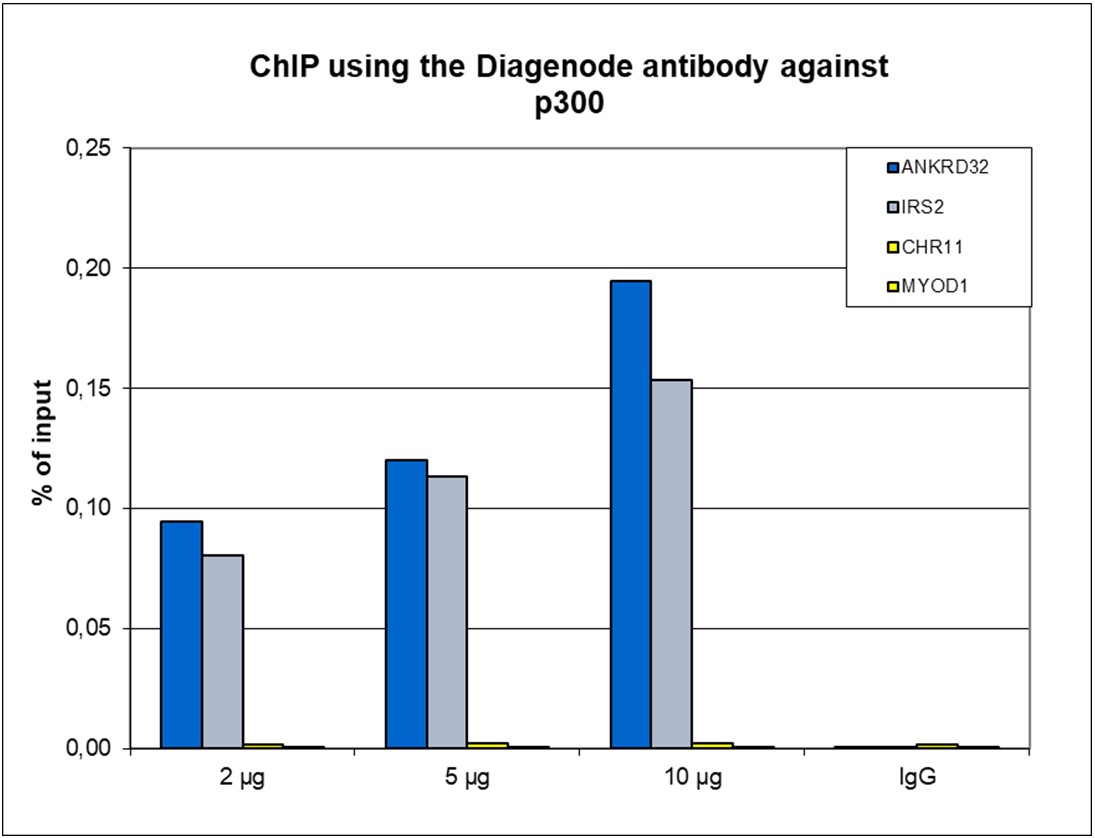
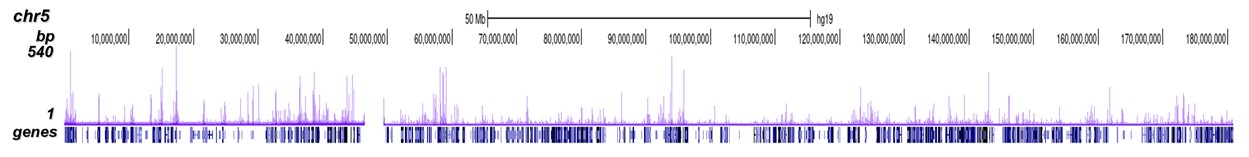
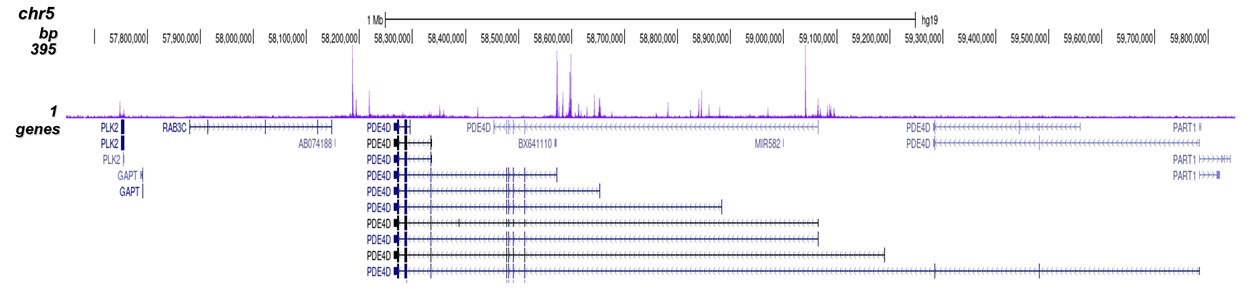
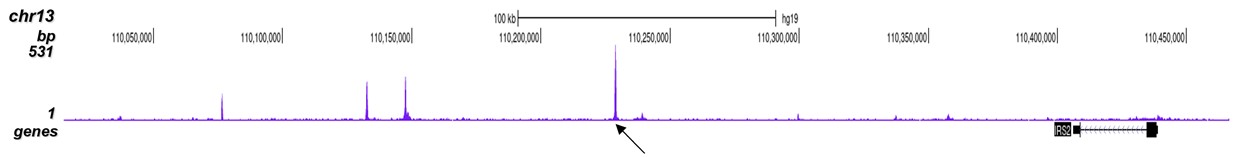
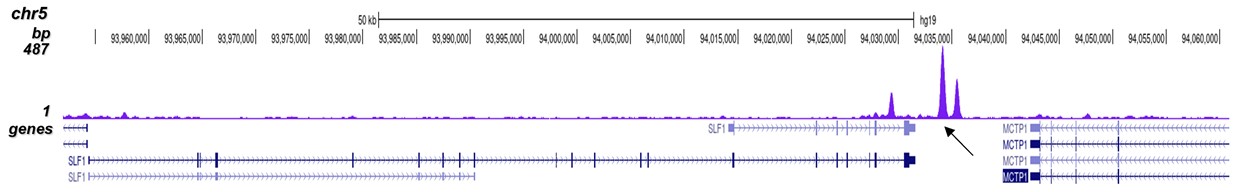
p300 (UniProt/Swiss-Prot entry Q09472) is a histone acetyltransferase that regulates transcription via chromatin remodelling. As such it is important for cell proliferation and differentiation. p300 is able to acetylate all four core histones in nucleosomes. Acetylation of histones is associated with transcriptional activation. p300 also acetylates non-histone proteins such as HDAC1 leading to its inactivation and modulation of transcription. It has also been identified as a co-activator of HIF1A (hypoxiainducible factor 1 alpha), and thus plays a role in the stimulation of hypoxia-induced genes such as VEGF. Defects in the p300 gene are a cause of Rubinstein-Taybi syndrome and may also play a role in epithelial cancer.