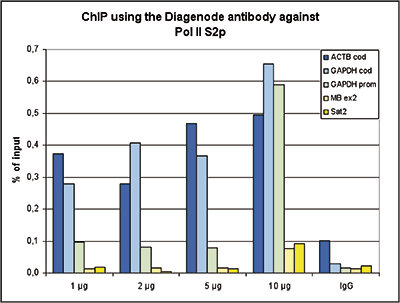
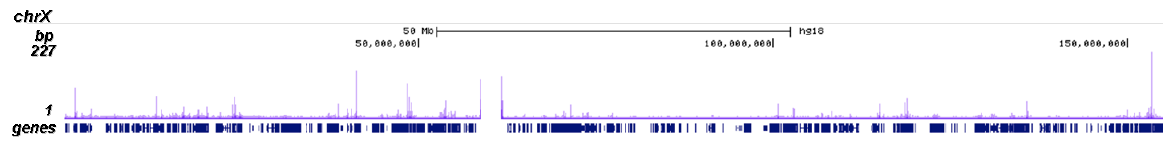
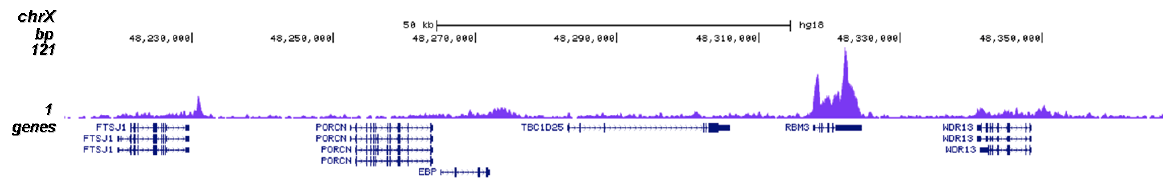
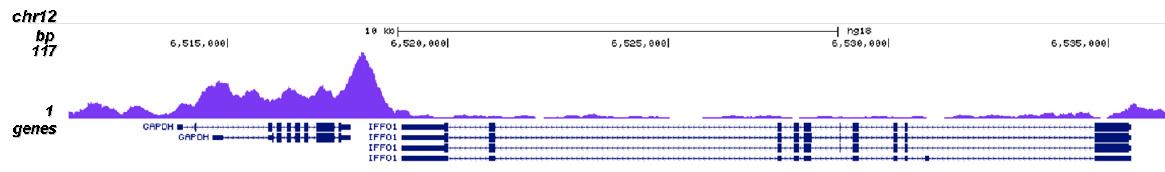
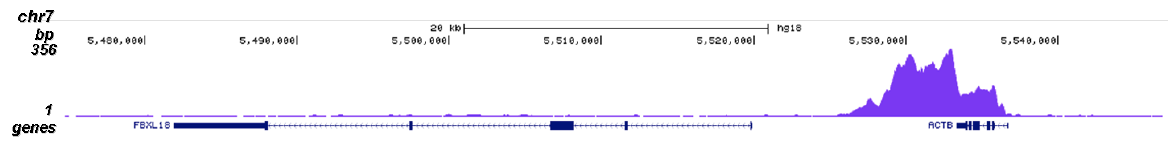
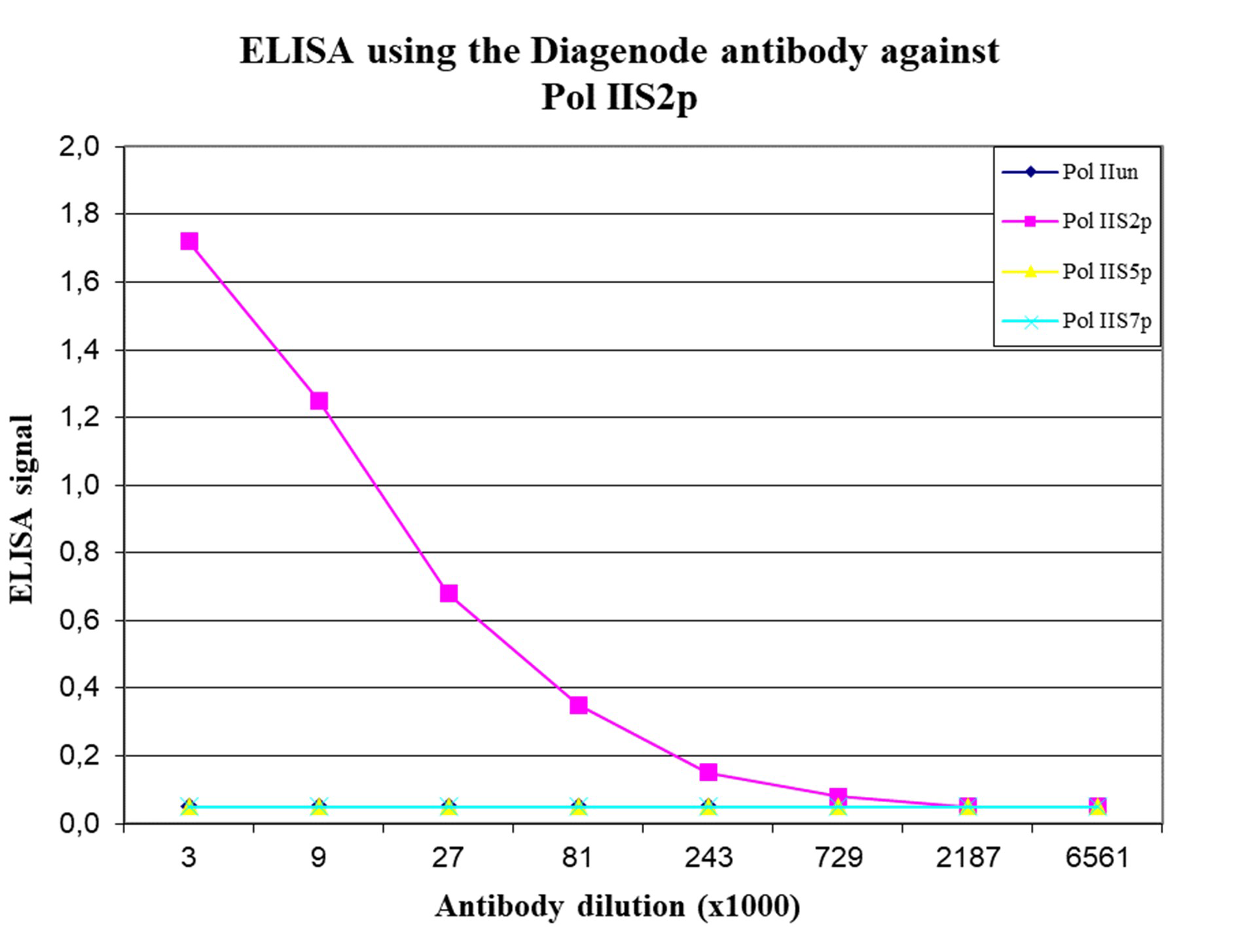
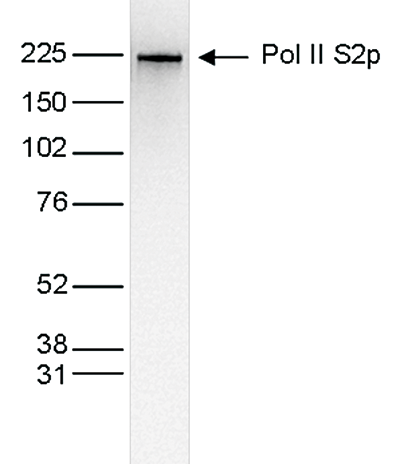
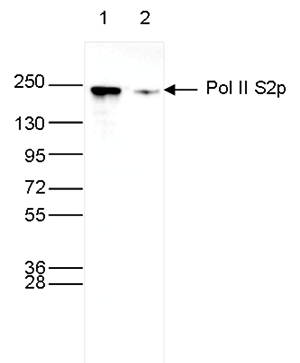
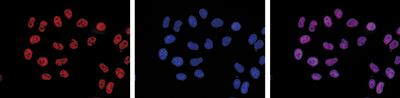
RNA polymerase II (pol II) is a key enzyme in the regulation and control of gene transcription. It is able to unwind the DNA double helix, synthesize RNA, and proofread the result. Pol II is a complex enzyme, consisting of 12 subunits, of which the B1 subunit (UniProt/Swiss-Prot entry P24928) is the largest. Together with the second largest subunit, B1 forms the catalytic core of the RNA polymerase II transcription machinery.