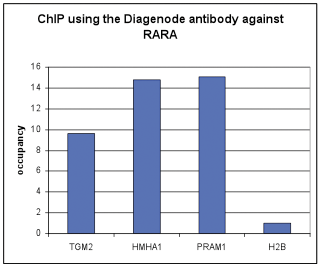
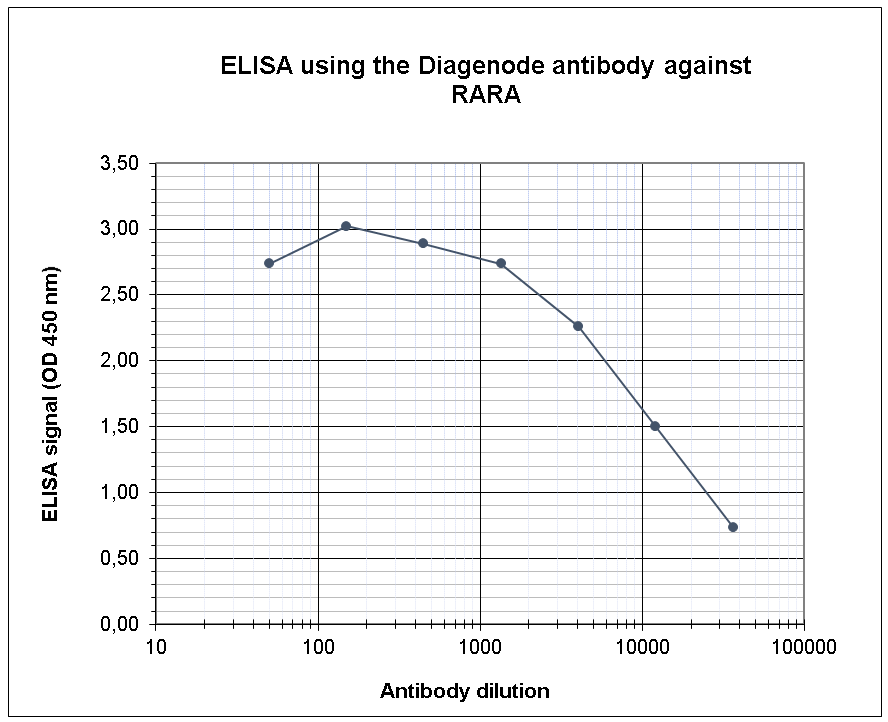
How to properly cite our product/service in your work We strongly recommend using this: RARA polyclonal antibody (Hologic Diagenode Cat# C15310155 Lot# A704-002). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Retinoic acid and ascorbate synergize to suppress myeloid leukemia via TET2 activation
Leesang, Tiffany E et al.
Enhancing ten-eleven translocation 2 (TET2) activity through genetic or pharmacologic approaches, such as ascorbate supplementation, can slow myeloid malignancy progression. However, ascorbate alone may be insufficient to fully activate TET2 in malignant cells due to pharmacokinetic constraints and the need for ch... |
ALDH1A1 promotes PARP inhibitor resistance by enhancing retinoic acidreceptor-mediated DNA polymerase θ expression.
Lavudi K. et al.
Poly (ADP-ribose) Polymerase (PARP) inhibitors (PARPi) have been approved for both frontline and recurrent setting in ovarian cancer with homologous recombination (HR) repair deficiency. However, more than 40\% of BRCA1/2-mutated ovarian cancer lack the initial response to PARPi treatment, and the majority of those ... |
Systems-biology analysis of rheumatoid arthritis fibroblast-likesynoviocytes implicates cell line-specific transcription factor function.
Ainsworth R. I. et al.
Rheumatoid arthritis (RA) is an immune-mediated disease affecting diarthrodial joints that remains an unmet medical need despite improved therapy. This limitation likely reflects the diversity of pathogenic pathways in RA, with individual patients demonstrating variable responses to targeted therapies. Better unders... |
Short-Chain Fatty Acids Calibrate RARα Activity RegulatingFood Sensitization
Yuan X. et al.
Gut-microbiota dysbiosis links to allergic diseases. The mechanism of the exacerbation of food allergy caused by gut-microbiota dysbiosis remains unknown. Regulation of retinoic acid receptor alpha (RARα) signaling is critical for gut immune homeostasis. Here we clarified that RARα in dendritic cells (DC... |
Retinoic Acid Receptor Alpha Represses a Th9 Transcriptional and Epigenomic Program to Reduce Allergic Pathology.
Schwartz DM, Farley TK, Richoz N, Yao C, Shih HY, Petermann F, Zhang Y, Sun HW, Hayes E, Mikami Y, Jiang K, Davis FP, Kanno Y, Milner JD, Siegel R, Laurence A, Meylan F, O'Shea JJ
CD4 T helper (Th) differentiation is regulated by diverse inputs, including the vitamin A metabolite retinoic acid (RA). RA acts through its receptor RARα to repress transcription of inflammatory cytokines, but is also essential for Th-mediated immunity, indicating complex effects of RA on Th specification and... |
Human retinoic acid-regulated CD161 regulatory T cells support wound repair in intestinal mucosa.
Povoleri GAM, Nova-Lamperti E, Scottà C, Fanelli G, Chen YC, Becker PD, Boardman D, Costantini B, Romano M, Pavlidis P, McGregor R, Pantazi E, Chauss D, Sun HW, Shih HY, Cousins DJ, Cooper N, Powell N, Kemper C, Pirooznia M, Laurence A, Kordasti S, Kazemi
Repair of tissue damaged during inflammatory processes is key to the return of local homeostasis and restoration of epithelial integrity. Here we describe CD161 regulatory T (T) cells as a distinct, highly suppressive population of T cells that mediate wound healing. These T cells were enriched in intestinal lamina ... |
Elevated Fibroblast growth factor 21 (FGF21) in obese, insulin resistant states is normalised by the synthetic retinoid Fenretinide in mice
Morrice N. et al.
Fibroblast growth factor 21 (FGF21) has emerged as an important beneficial regulator of glucose and lipid homeostasis but its levels are also abnormally increased in insulin-resistant states in rodents and humans. The synthetic retinoid Fenretinide inhibits obesity and improves glucose homeostasis in mice and has pl... |
The oncofusion protein FUS-ERG targets key hematopoietic regulators and modulates the all-trans retinoic acid signaling pathway in t(16;21) acute myeloid leukemia.
Sotoca AM1, Prange KH1, Reijnders B1, Mandoli A1, Nguyen LN1, Stunnenberg HG1, Martens JH
The ETS transcription factor ERG has been implicated as a major regulator of both normal and aberrant hematopoiesis. In acute myeloid leukemias harboring t(16;21), ERG function is deregulated due to a fusion with FUS/TLS resulting in the expression of a FUS-ERG oncofusion protein. How this oncofusion protein deregul... |
Retinoic acid is essential for Th1 cell lineage stability and prevents transition to a Th17 cell program.
Brown CC, Esterhazy D, Sarde A, London M, Pullabhatla V, Osma-Garcia I, Al-Bader R, Ortiz C, Elgueta R, Arno M, de Rinaldis E, Mucida D, Lord GM, Noelle RJ
CD4(+) T cells differentiate into phenotypically distinct T helper cells upon antigenic stimulation. Regulation of plasticity between these CD4(+) T-cell lineages is critical for immune homeostasis and prevention of autoimmune disease. However, the factors that regulate lineage stability are largely unknown. Here we... |
PML-RARalpha/RXR Alters the Epigenetic Landscape in Acute Promyelocytic Leukemia.
Martens JH, Brinkman AB, Simmer F, Francoijs KJ, Nebbioso A, Ferrara F, Altucci L, Stunnenberg HG
Many different molecular mechanisms have been associated with PML-RARalpha-dependent transformation of hematopoietic progenitors. Here, we identified high confidence PML-RARalpha binding sites in an acute promyelocytic leukemia (APL) cell line and in two APL primary blasts. We found colocalization of PML-RARalpha wi... |