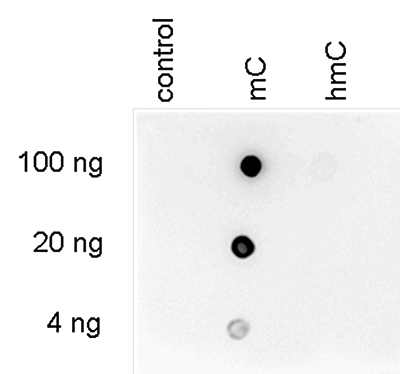
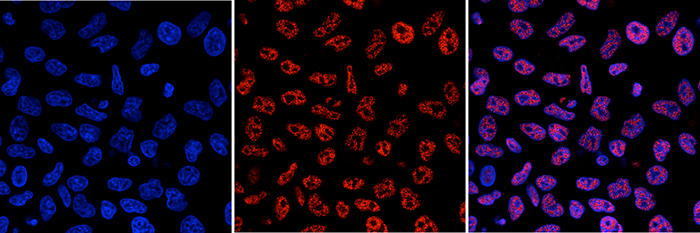
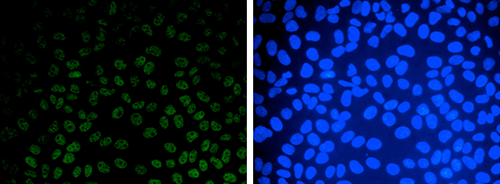
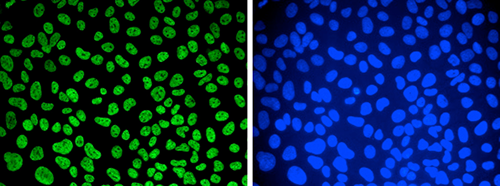
Developmental mRNA mC landscape and regulatory innovations of massivemC modification of maternal mRNAs in animals.
Liu J. et al.
mC is one of the longest-known RNA modifications, however, its developmental dynamics, functions, and evolution in mRNAs remain largely unknown. Here, we generate quantitative mRNA mC maps at different stages of development in 6 vertebrate and invertebrate species and find convergent and unexpected massive methylati... |
Transcriptome-wide distribution and function of RNA hydroxymethylcytosine
Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, Wetzel C, Kreher J, Soin R, Creppe C, Limbach PA, Gueydan C, Kruys V, Brehm A, Minakhina S, Defrance M, Steward R, Fuks F.
Hydroxymethylcytosine, well described in DNA, occurs also in RNA. Here, we show that hydroxymethylcytosine preferentially marks polyadenylated RNAs and is deposited by Tet in Drosophila. We map the transcriptome-wide hydroxymethylation landscape, revealing hydroxymethylcytosine in the transcripts of many genes, nota... |
RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine.
Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, Wetzel C, Kreher J, Soin R, Creppe C, Limbach PA, Gueydan C, Kruys V, Brehm A, Minakhina S, Defrance M, Steward R, Fuks F
Hydroxymethylcytosine, well described in DNA, occurs also in RNA. Here, we show that hydroxymethylcytosine preferentially marks polyadenylated RNAs and is deposited by Tet in Drosophila. We map the transcriptome-wide hydroxymethylation landscape, revealing hydroxymethylcytosine in the transcripts of many genes, nota... |