The Diagenode ChIPmentation technology has been tested on histone marks and compared to available datasets from the ENCODE project (Figure 1). ChIPmentation generated high quality data with low background. In addition, more than 99% of the top 40% peaks obtained with auto-ChIPmentation overlap with ENCODE datasets, which shows that ChIP-seq data obtained with ChIPmentation are highly reliable.
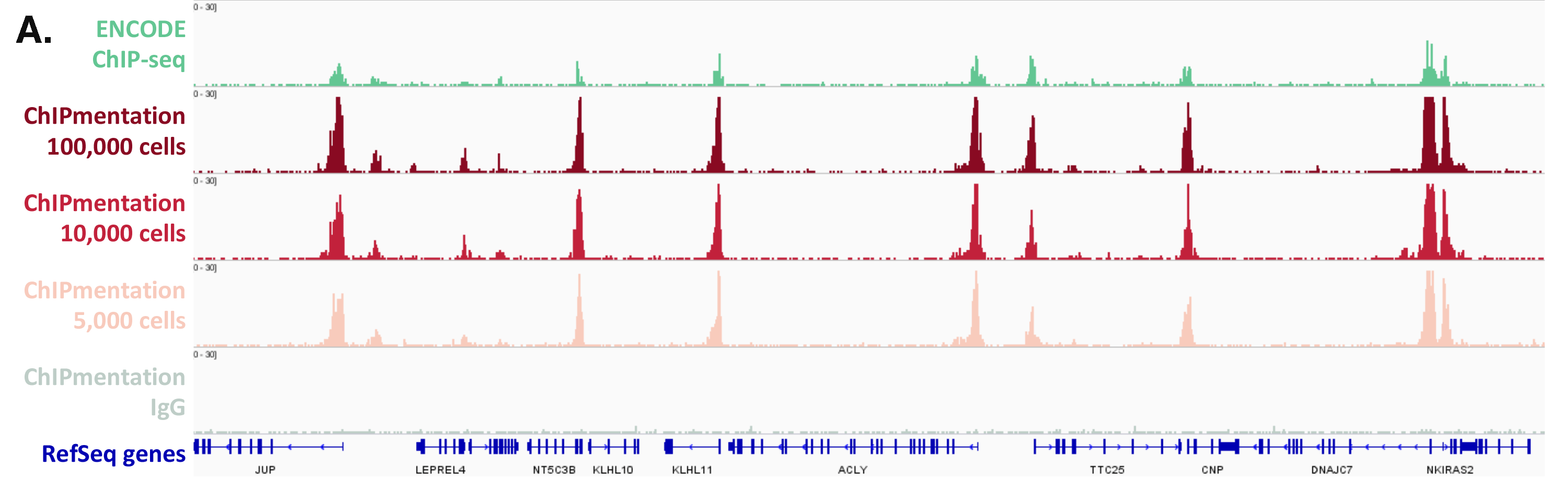
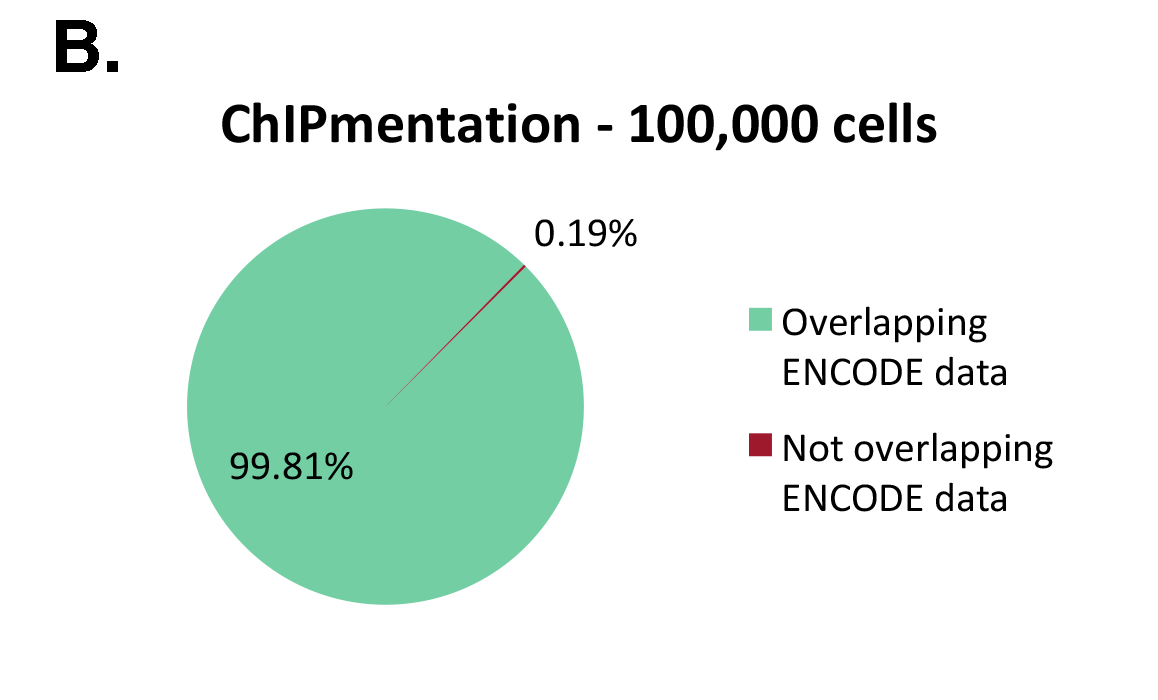
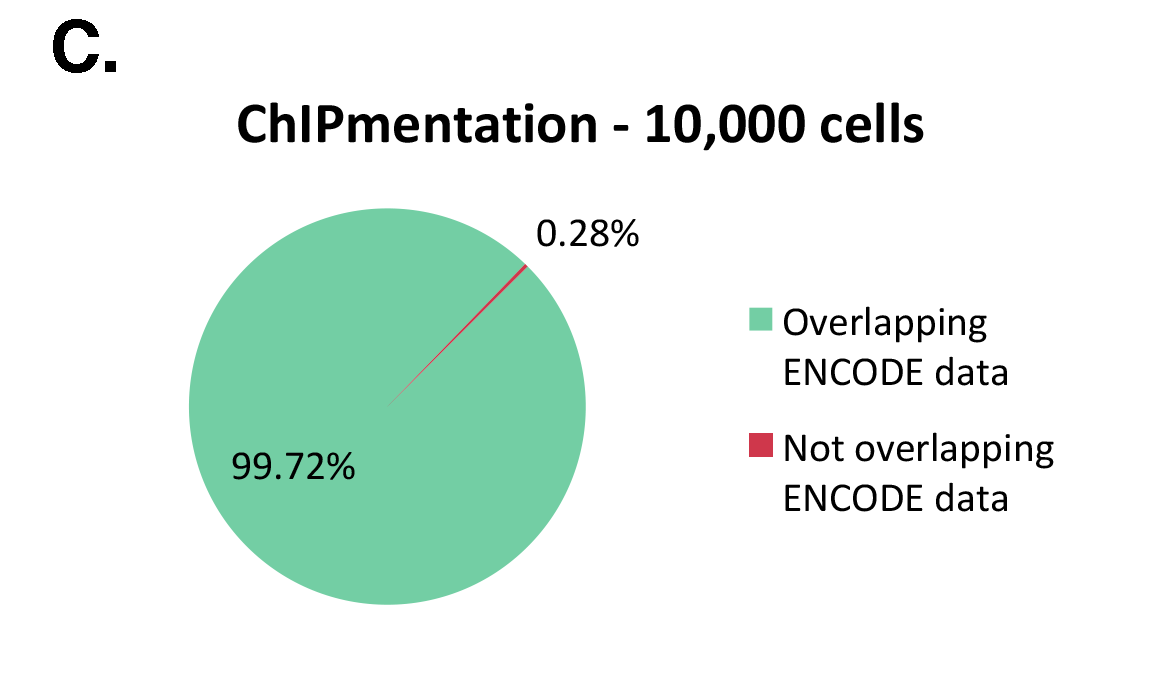
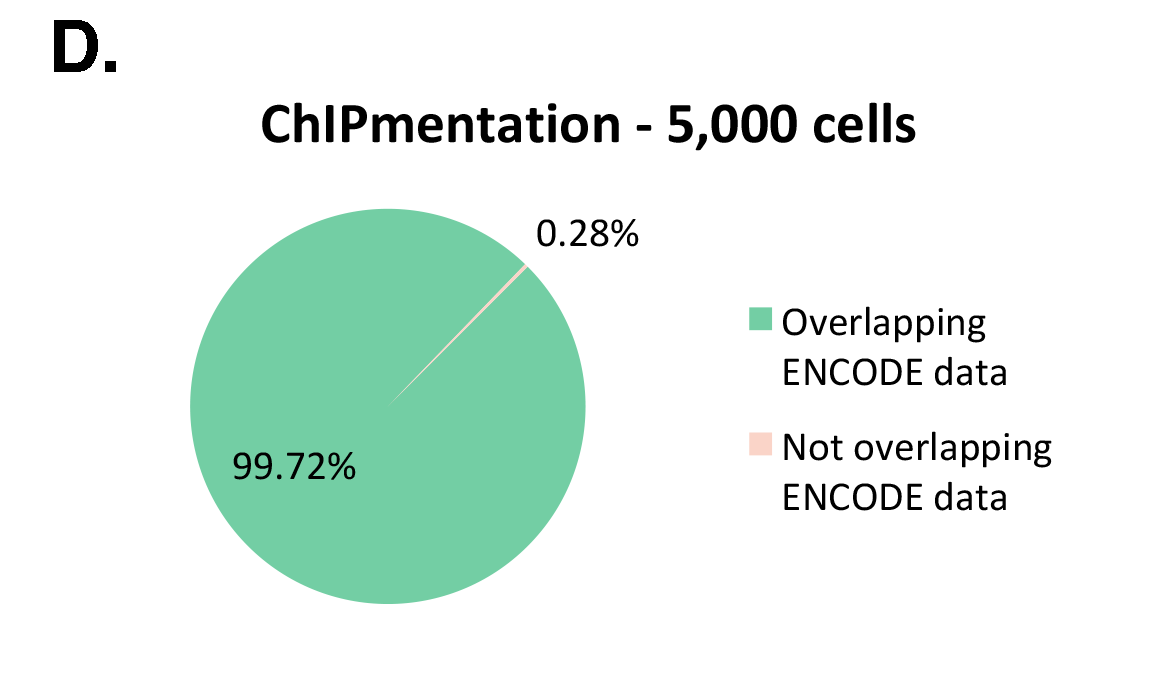
Figure 1: ChIPmentation sequencing results obtained from decreasing starting amounts of cells.
Chromatin preparation has been performed on 7 M K562 cells using the ChIPmentation Kit for Histones (Cat. no. C01011009) and 24 SI for ChIPmentation (Cat. No. C01011031). Diluted chromatin from 100.000, 10.000 and 5.000 cells was used for the immunoprecipitation with the Diagenode antibody targeting H3K4me3 (Cat. no. C15410003). A. Distribution of the ChIPmentation readsets in a representative region of the genome. B., C. and D. Comparison of the top 40% peaks from 100.000 (B.), 10,000 (C.) and 5.000 (D.) cells with ENCODE dataset.
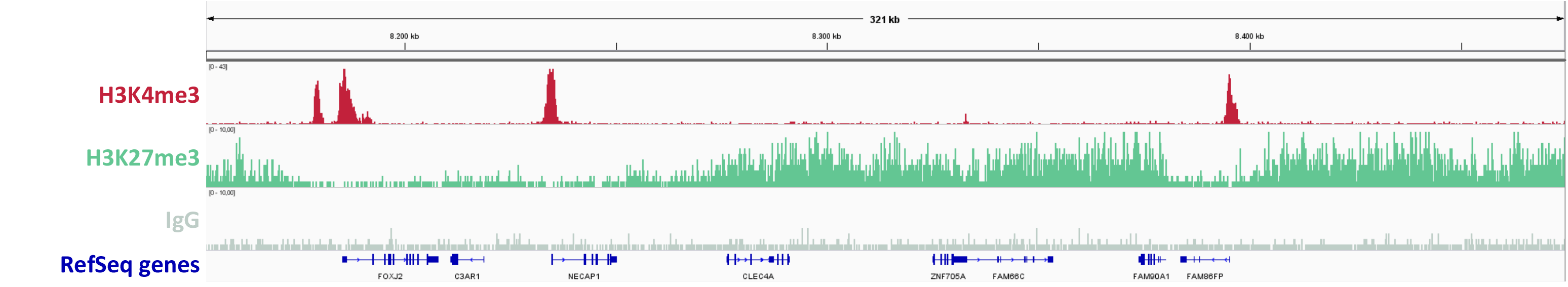
Figure 2: ChIPmentation sequencing results.
Chromatin preparation has been performed on 7 M HeLa cells using the ChIPmentation Kit for Histones and 24 SI for ChIPmentation. Diluted chromatin from 100.000 cells was used for the immunoprecipitation with the Diagenode antibody targeting H3K4me3 (Cat. no. C15410003) and H3K27me3 (Cat. no. C15410195) and IgG (Cat. no. C15410206).