How to properly cite our product/service in your work We strongly recommend using this: Ty1 monoclonal antibody (Hologic Diagenode Cat# C15200054 Lot# 008). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Profiling BRCA1-BRCT interactions and their functional relevance at amino acid resolution
Mangkusaputra, Venda et al.
BRCA1 (breast cancer-associated protein 1) plays a central role in homologous recombination (HR) through interactions with multiple proteins across its various domains. The C-terminal BRCT domains bind HR regulators such as ABRAXAS1, CtIP, and BRIP1, each contributing to distinct, sometimes opposing, functions. ... |
Somatic mutations in TBX3 promote hepatic clonal expansion by accelerating VLDL secretion
Mannino, Gregory et al.
Somatic mutations that increase clone fitness or resist disease are positively selected, but the impact of these mutations on organismal health remains unclear. We previously showed that Tbx3 deletion increases hepatocyte fitness within fatty livers. Here, we detected TBX3 somatic mutations in patients with metabo... |
The small inhibitor WM-1119 effectively targets KAT6A-rearranged AML, but not KMT2A-rearranged AML, despite shared KAT6 genetic dependency
Mathew Sheridan et al.
Background
The epigenetic factors KAT6A (MOZ/MYST3) and KMT2A (MLL/MLL1) interact in normal hematopoiesis to regulate progenitors’ self-renewal. Both proteins are recurrently translocated in AML, leading to impairment of critical differentiation pathways in these malignant cells. We evaluated the potential of... |
Zfp296 knockout enhances chromatin accessibility and induces a uniquestate of pluripotency in embryonic stem cells.
Miyazaki S. et al.
The Zfp296 gene encodes a zinc finger-type protein. Its expression is high in mouse embryonic stem cells (ESCs) but rapidly decreases following differentiation. Zfp296-knockout (KO) ESCs grew as flat colonies, which were reverted to rounded colonies by exogenous expression of Zfp296. KO ESCs could not form teratomas... |
Net39 protects muscle nuclei from mechanical stress during thepathogenesis of Emery-Dreifuss muscular dystrophy.
Zhang Y. et al.
Mutations in genes encoding nuclear envelope proteins lead to diseases known as nuclear envelopathies, characterized by skeletal muscle and heart abnormalities, such as Emery-Dreifuss muscular dystrophy (EDMD). The tissue-specific role of the nuclear envelope in the etiology of these diseases has not been extensivel... |
Distinct effects on the secretion of MTRAP and AMA1 in Plasmodiumyoelii following deletion of acylated pleckstrin homology domain-containingprotein.
Chaiyawong Nattawat et al.
Plasmodium, the causative agents of malaria, are obligate intracellular organisms. In humans, pathogenesis is caused by the blood stage parasite, which multiplies within erythrocytes, thus erythrocyte invasion is an essential developmental step. Merozoite form parasites released into the blood stream coordinately se... |
Mod(mdg4) variants repress telomeric retrotransposon HeT-A byblocking subtelomeric enhancers
Takeuchi Chikara et al.
Telomeres in Drosophila are composed of sequential non-LTR retrotransposons: HeT-A, TART , and TAHRE . Although they are repressed by the piRNA pathway in the germline, how these retrotransposons are regulated in somatic cells is poorly understood. Here, we show that specific splice variants of Mod(mdg4) repress HeT... |
Identification of a PadR-type regulator essential for intracellularpathogenesis of
McMillan Ian A. et al.
Burkholderia pseudomallei (Bp) is the causative agent of melioidosis, a disease endemic to the tropics. Melioidosis manifests in various ways ranging from acute skin lesions to pneumonia and, in rare cases, infection of the central nervous system. Bp is a facultative intracellular pathogen and it can infect various ... |
MEAF6 is essential for cell proliferation and plays a role in theassembly of KAT7 complexes.
Matsuura, Kumi and Tani, Naoki and Usuki, Shingo and Torikai-Nishikawa,Satomi and Okano, Masaki and Niwa, Hitoshi
Myst family genes encode lysine acetyltransferases that mainly mediate histone acetylation to control transcription, DNA replication and DNA damage response. They form tetrameric complexes with PHD-finger proteins (Brpfs or Jades) and small non-catalytic subunits Ing4/5 and Meaf6. Although all the components of the ... |
Dual ARID1A/ARID1B loss leads to rapid carcinogenesis and disruptiveredistribution of BAF complexes
Wang, Zixi and Chen, Kenian and Jia, Yuemeng and Chuang, Jen-Chieh and Sun,Xuxu and Lin, Yu-Hsuan and Celen, Cemre and Li, Lin and Huang, Fang andLiu, Xin and Castrillon, Diego H. and Wang, Tao and Zhu, Hao
SWI/SNF chromatin remodelers play critical roles in development and cancer. The causal links between SWI/SNF complex disassembly and carcinogenesis are obscured by redundancy between paralogous components. Canonical BAF (cBAF)-specific paralogs ARID1A and ARID1B are synthetic lethal in some contexts, but simultaneou... |
miR-155 inhibits mitophagy through suppression of BAG5, a partner protein of PINK1.
Tsujimoto T, Mori T, Houri K, Onodera Y, Takehara T, Shigi K, Nakao S, Teramura T, Fukuda K
Removal of dysfunctional mitochondria is essential step to maintain normal cell physiology, and selective autophagy in mitochondria, called mitophagy, plays a critical role in quality control of mitochondria. While in several diseases and aging, disturbed mitophagy has been observed. In stem cells, accumulation of d... |
Twist2 amplification in rhabdomyosarcoma represses myogenesis and promotes oncogenesis by redirecting MyoD DNA binding.
Li S, Chen K, Zhang Y, Barnes SD, Jaichander P, Zheng Y, Hassan M, Malladi VS, Skapek SX, Xu L, Bassel-Duby R, Olson EN, Liu N
Rhabdomyosarcoma (RMS) is an aggressive pediatric cancer composed of myoblast-like cells. Recently, we discovered a unique muscle progenitor marked by the expression of the Twist2 transcription factor. Genomic analyses of 258 RMS patient tumors uncovered prevalent copy number amplification events and increased expre... |
Cardiac Reprogramming Factors Synergistically Activate Genome-wide Cardiogenic Stage-Specific Enhancers.
Hashimoto H, Wang Z, Garry GA, Malladi VS, Botten GA, Ye W, Zhou H, Osterwalder M, Dickel DE, Visel A, Liu N, Bassel-Duby R, Olson EN
The cardiogenic transcription factors (TFs) Mef2c, Gata4, and Tbx5 can directly reprogram fibroblasts to induced cardiac-like myocytes (iCLMs), presenting a potential source of cells for cardiac repair. While activity of these TFs is enhanced by Hand2 and Akt1, their genomic targets and interactions during reprogram... |
Plasmodium falciparum gametocyte-infected erythrocytes do not adhere to human primary erythroblasts.
Neveu G, Dupuy F, Ladli M, Barbieri D, Naissant B, Richard C, Martins RM, Lopez-Rubio JJ, Bachmann A, Verdier F, Lavazec C
Plasmodium falciparum gametocytes, the sexual stages responsible for malaria parasite transmission, develop in the human bone marrow parenchyma in proximity to the erythroblastic islands. Yet, mechanisms underlying gametocytes interactions with these islands are unknown. Here, we have investigated whether gametocyte... |
Novel dual regulators of Pseudomonas aeruginosa essential for productive biofilms and virulence.
Heacock-Kang Y, Zarzycki-Siek J, Sun Z, Poonsuk K, Bluhm AP, Cabanas D, Fogen D, McMillan IA, Chuanchuen R, Hoang TT
Gene regulation network in Pseudomonas aeruginosa is complex. With a relatively large genome (6.2 Mb), there is a significant portion of genes that are proven or predicted to be transcriptional regulators. Many of these regulators have been shown to play important roles in biofilm formation and maintenance. In this ... |
Cockayne's Syndrome A and B Proteins Regulate Transcription Arrest after Genotoxic Stress by Promoting ATF3 Degradation
Epanchintsev A. et al.
Cockayne syndrome (CS) is caused by mutations in CSA and CSB. The CSA and CSB proteins have been linked to both promoting transcription-coupled repair and restoring transcription following DNA damage. We show that UV stress arrests transcription of approximately 70% of genes in CSA- or CSB-deficient cells due to the... |
Gene regulatory networks in neural cell fate acquisition from genome-wide chromatin association of Geminin and Zic1
Sankar S. et al.
Neural cell fate acquisition is mediated by transcription factors expressed in nascent neuroectoderm, including Geminin and members of the Zic transcription factor family. However, regulatory networks through which this occurs are not well defined. Here, we identified Geminin-associated chromatin locations in embryo... |
Severe muscle wasting and denervation in mice lacking the RNA-binding protein ZFP106
Anderson DM et al.
Innervation of skeletal muscle by motor neurons occurs through the neuromuscular junction, a cholinergic synapse essential for normal muscle growth and function. Defects in nerve-muscle signaling cause a variety of neuromuscular disorders with features of ataxia, paralysis, skeletal muscle wasting, and degeneration.... |
Converging disease genes in ICF syndrome: ZBTB24 controls expression of CDCA7 in mammals
Wu H et al.
For genetically heterogeneous diseases a better understanding of how the underlying gene defects are functionally interconnected will be important for dissecting disease etiology. The Immunodeficiency, Centromeric instability, Facial anomalies (ICF) syndrome is a chromatin disorder characterized by mutations in DNMT... |
Osr1 Interacts Synergistically with Wt1 to Regulate Kidney Organogenesis
Xu J et al.
Renal hypoplasia is a common cause of pediatric renal failure and several adult-onset diseases. Recent studies have associated a variant of the OSR1 gene with reduction of newborn kidney size and function in heterozygotes and neonatal lethality with kidney defects in homozygotes. How OSR1 regulates kidney developmen... |
Zbtb20 promotes astrocytogenesis during neocortical development
Nagao M, Ogata T, Sawada Y, Gotoh Y
Multipotent neural precursor cells (NPCs) generate astrocytes at late stages of mammalian neocortical development. Many signalling pathways that regulate astrocytogenesis directly induce the expression of GFAP, a marker of terminally differentiated astrocytes. However, astrocyte specification occurs before GFAP expr... |
Expression of the MOZ-TIF2 oncoprotein in mice represses senescence.
Largeot A, Perez-Campo FM, Marinopoulou E, Lie-A-Ling M, Kouskoff V, Lacaud G.
The MOZ-TIF2 translocation, that fuses MOZ (Monocytic Leukemia Zinc finger protein) histone acetyltransferase (HAT) with the nuclear co-activator TIF2, is associated the development of Acute Myeloid Leukemia. We recently showed that in the absence of MOZ HAT activity, p16INK4atranscriptional levels are sig... |
The Cytoplasmic Region of Plasmodium falciparum SURFIN4.2 Is Required for Transport from Maurer’s Clefts to the Red Blood Cell Surface
Kagaya W et al.
Background: Plasmodium, the causative agent of malaria, exports many proteins to the surface of the infected red blood cell (iRBC) in order to modify it toward a structure more suitable for parasite development and survival. One such exported protein, SURFIN4.2, from the parasite of human malignant malaria, P. fal... |
TAL effector-mediated genome visualization (TGV).
Miyanari Y
The three-dimensional remodeling of chromatin within nucleus is being recognized as determinant for genome regulation. Recent technological advances in live imaging of chromosome loci begun to explore the biological roles of the movement of the chromatin within the nucleus. To facilitate better understanding of the ... |
Live visualization of chromatin dynamics with fluorescent TALEs.
Miyanari Y, Ziegler-Birling C, Torres-Padilla ME
The spatiotemporal organization of genomes in the nucleus is an emerging key player to regulate genome function. Live imaging of nuclear organization dynamics would be a breakthrough toward uncovering the functional relevance and mechanisms regulating genome architecture. Here, we used transcription activator-like e... |
Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells.
Morales Torres C, Laugesen A, Helin K
Embryonic development requires chromatin remodeling for dynamic regulation of gene expression patterns to ensure silencing of pluripotent transcription factors and activation of developmental regulators. Demethylation of H3K27me3 by the histone demethylases Utx and Jmjd3 is important for the activation of lineage ch... |
The N-terminal segment of Plasmodium falciparum SURFIN(4.1) is required for its trafficking to the red blood cell cytosol through the endoplasmic reticulum.
Zhu X, Yahata K, Alexandre JS, Tsuboi T, Kaneko O
Plasmodium falciparum SURFIN is a type I transmembrane protein that shares domains with molecules expressed on the surface of the red blood cells (RBCs) infected with a variety of malaria parasite species, such as P. falciparum PfEMP1, P. vivax VIR proteins, and P. knowlesi SICAvar. Thus, understanding the export me... |
The Human EKC/KEOPS Complex Is Recruited to Cullin2 Ubiquitin Ligases by the Human Tumour Antigen PRAME.
Costessi A, Mahrour N, Sharma V, Stunnenberg R, Stoel MA, Tijchon E, Conaway JW, Conaway RC, Stunnenberg HG
The human tumour antigen PRAME (preferentially expressed antigen in melanoma) is frequently overexpressed during oncogenesis, and high PRAME levels are associated with poor clinical outcome in a variety of cancers. However, the molecular pathways in which PRAME is implicated are not well understood. We recently char... |
An essential novel component of the non-canonical mitochondrial outer membrane protein import system of trypanosomatids.
Pusnik M, Mani J, Schmidt O, Niemann M, Oeljeklaus S, Schnarwiler F, Warscheid B, Lithgow T, Meisinger C, Schneider A.
The mitochondrial outer membrane protein Tom40 is the general entry gate for imported proteins in essentially all eukaryotes. Trypanosomatids however lack Tom40 and use instead a protein termed the archaic translocase of the outer mitochondrial membrane (ATOM). Here we report the discovery of pATOM36, a novel essent... |
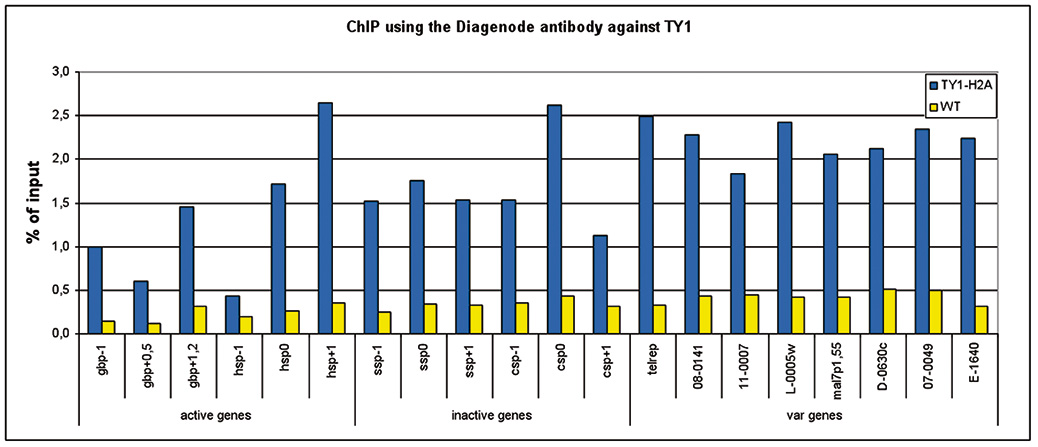
Selective autophagy regulates insertional mutagenesis by the Ty1 retrotransposon in Saccharomyces cerevisiae.
Suzuki K, Morimoto M, Kondo C, Ohsumi Y
Macroautophagy (autophagy) is a bulk degradation system for cytoplasmic components and is ubiquitously found in eukaryotic cells. Autophagy is induced under starvation conditions and plays a cytoprotective role by degrading unwanted cytoplasmic materials. The Ty1 transposon, a member of the Ty1/copia superfamily, is... |
The tumour antigen PRAME is a subunit of a Cul2 ubiquitin ligase and associates with active NFY promoters.
Costessi A, Mahrour N, Tijchon E, Stunnenberg R, Stoel MA, Jansen PW, Sela D, Martin-Brown S, Washburn MP, Florens L, Conaway JW, Conaway RC, Stunnenberg HG
The human tumour antigen PRAME (preferentially expressed antigen of melanoma) is frequently overexpressed in tumours. High PRAME levels correlate with poor clinical outcome of several cancers, but the mechanisms by which PRAME could be involved in tumourigenesis remain largely elusive. We applied protein-complex pur... |
Recombineering, transfection, Western, IP and ChIP methods for protein tagging via gene targeting or BAC transgenesis.
Hofemeister H, Ciotta G, Fu J, Seibert PM, Schulz A, Maresca M, Sarov M, Anastassiadis K, Stewart AF
Protein tagging offers many advantages for proteomic and regulomic research. Ideally, protein tagging is equivalent to having a high affinity antibody for every chosen protein. However, these advantages are compromised if the tagged protein is overexpressed, which is usually the case from cDNA expression vectors. Ph... |