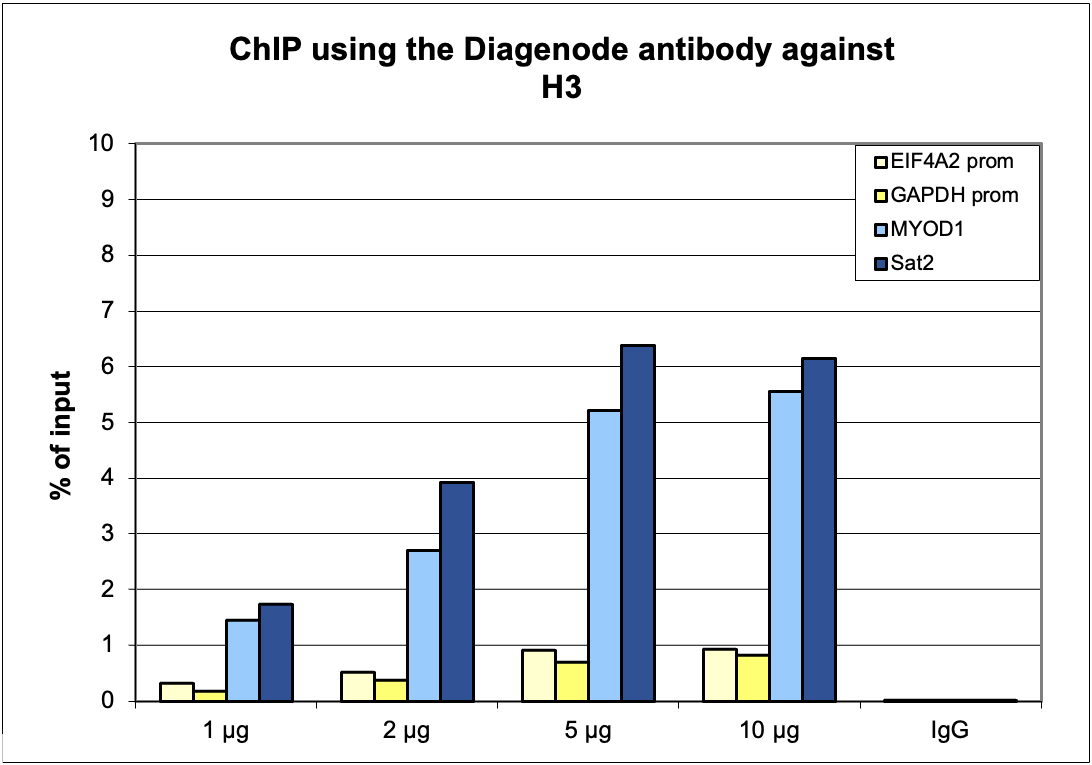
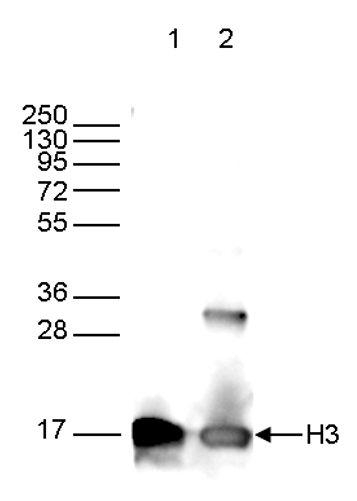
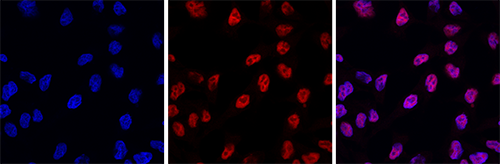
How to properly cite our product/service in your work We strongly recommend using this: H3pan Antibody 1B1B2 (Hologic Diagenode Cat# C15200011 Lot# 003). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Temporal modification of H3K9/14ac and H3K4me3 histone marksmediates mechano-responsive gene expression during the accommodationprocess in poplar
Ghosh R. et al.
Plants can attenuate their molecular response to repetitive mechanical stimulation as a function of their mechanical history. For instance, a single bending of stem is sufficient to attenuate the gene expression in poplar plants to the subsequent mechanical stimulation, and the state of desensitization can last for ... |
Trichoderma root colonization triggers epigenetic changes in jasmonic andsalicylic acid pathway-related genes.
Agostini R. B. et al.
Beneficial interactions between plant-roots and Trichoderma spp. lead to a local and systemic enhancement of the plant immune system through a mechanism known as priming of defenses. In recent reports, we outlined a repertoire of genes and proteins differentially regulated in distant tissues of maize plants previous... |
Nox4 promotes endothelial differentiation through chromatin remodeling.
Hahner F. et al.
RATIONALE: Nox4 is a constitutively active NADPH oxidase that constantly produces low levels of HO. Thereby, Nox4 contributes to cell homeostasis and long-term processes, such as differentiation. The high expression of Nox4 seen in endothelial cells contrasts with the low abundance of Nox4 in stem cells, which are a... |
Single amino-acid mutation in a Drosoph ila melanogaster ribosomalprotein: An insight in uL11 transcriptional activity.
Grunchec H. et al.
The ribosomal protein uL11 is located at the basis of the ribosome P-stalk and plays a paramount role in translational efficiency. In addition, no mutant for uL11 is available suggesting that this gene is haplo-insufficient as many other Ribosomal Protein Genes (RPGs). We have previously shown that overexpression of... |
Activated Histone Acetyltransferase p300/CBP-Related SignallingPathways Mediate Up-Regulation of NADPH Oxidase,Inflammation, and Fibrosis in Diabetic Kidney
Alexandra-Gela Lazar et al.
Accumulating evidence implicates the histone acetylation-based epigenetic mechanisms in the pathoetiology of diabetes-associated micro-/macrovascular complications. Diabetic kidney disease (DKD) is a progressive chronic inflammatory microvascular disorder ultimately leading to glomerulosclerosis and kidney failure. ... |
ZNF354C is a transcriptional repressor that inhibits endothelialangiogenic sprouting.
Oo, James A and Irmer, Barnabas and Günther, Stefan and Warwick, Timothyand Pálfi, Katalin and Izquierdo Ponce, Judit and Teichmann, Tom andPflüger-Müller, Beatrice and Gilsbach, Ralf and Brandes, Ralf P andLeisegang, Matthias S
Zinc finger proteins (ZNF) are a large group of transcription factors with diverse functions. We recently discovered that endothelial cells harbour a specific mechanism to limit the action of ZNF354C, whose function in endothelial cells is unknown. Given that ZNF354C has so far only been studied in bone and tum... |
Revisiting promyelocytic leukemia protein targeting by human cytomegalovirus immediate-early protein 1.
Paulus C, Harwardt T, Walter B, Marxreiter A, Zenger M, Reuschel E, Nevels MM
Promyelocytic leukemia (PML) bodies are nuclear organelles implicated in intrinsic and innate antiviral defense. The eponymous PML proteins, central to the self-organization of PML bodies, and other restriction factors found in these organelles are common targets of viral antagonism. The 72-kDa immediate-early prote... |
Widespread loss of the silencing epigenetic mark H3K9me3 in astrocytes and neurons along with hippocampal-dependent cognitive impairment in C9orf72 BAC transgenic mice.
Jury N, Abarzua S, Diaz I, Guerra MV, Ampuero E, Cubillos P, Martinez P, Herrera-Soto A, Arredondo C, Rojas F, Manterola M, Rojas A, Montecino M, Varela-Nallar L, van Zundert B
BACKGROUND: Hexanucleotide repeat expansions of the GC motif in a non-coding region of the C9ORF72 gene are the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Tissues from C9ALS/FTD patients and from mouse models of ALS show RNA foci, dipeptide-repeat proteins, an... |
Class I TCP transcription factors target the gibberellin biosynthesis gene GA20ox1 and the growth promoting genes HBI1 and PRE6 during thermomorphogenic growth in Arabidopsis.
Ferrero LV, Viola IL, Ariel FD, Gonzalez DH
Plants respond to a rise in ambient temperature by increasing the growth of petioles and hypocotyls. In this work, we show that Arabidopsis thaliana class I TEOSINTE BRANCHED 1, CYCLOIDEA, PCF (TCP) transcription factors TCP14 and TCP15 are required for optimal petiole and hypocotyl elongation under high ambient tem... |
Comprehensive Analysis of Chromatin States in Atypical Teratoid/Rhabdoid Tumor Identifies Diverging Roles for SWI/SNF and Polycomb in Gene Regulation.
Erkek S, Johann PD, Finetti MA, Drosos Y, Chou HC, Zapatka M, Sturm D, Jones DTW, Korshunov A, Rhyzova M, Wolf S, Mallm JP, Beck K, Witt O, Kulozik AE, Frühwald MC, Northcott PA, Korbel JO, Lichter P, Eils R, Gajjar A, Roberts CWM, Williamson D, Hasselbla
Biallelic inactivation of SMARCB1, encoding a member of the SWI/SNF chromatin remodeling complex, is the hallmark genetic aberration of atypical teratoid rhabdoid tumors (ATRT). Here, we report how loss of SMARCB1 affects the epigenome in these tumors. Using chromatin immunoprecipitation sequencing (ChIP-seq) on pri... |
Epigenetic regulation of vascular NADPH oxidase expression and reactive oxygen species production by histone deacetylase-dependent mechanisms in experimental diabetes.
Manea SA, Antonescu ML, Fenyo IM, Raicu M, Simionescu M, Manea A
Reactive oxygen species (ROS) generated by up-regulated NADPH oxidase (Nox) contribute to structural-functional alterations of the vascular wall in diabetes. Epigenetic mechanisms, such as histone acetylation, emerged as important regulators of gene expression in cardiovascular disorders. Since their role in diabete... |
The histone demethylase Phf2 acts as a molecular checkpoint to prevent NAFLD progression during obesity.
Bricambert J, Alves-Guerra MC, Esteves P, Prip-Buus C, Bertrand-Michel J, Guillou H, Chang CJ, Vander Wal MN, Canonne-Hergaux F, Mathurin P, Raverdy V, Pattou F, Girard J, Postic C, Dentin R
Aberrant histone methylation profile is reported to correlate with the development and progression of NAFLD during obesity. However, the identification of specific epigenetic modifiers involved in this process remains poorly understood. Here, we identify the histone demethylase Plant Homeodomain Finger 2 (Phf2) as a... |
Expression of the Parkinson's Disease-Associated Gene Alpha-Synuclein is Regulated by the Neuronal Cell Fate Determinant TRIM32
Pavlou MA et al.
Alpha-synuclein is an abundant neuronal protein which has been associated with physiological processes like synaptic function, neurogenesis, and neuronal differentiation but also with pathological neurodegeneration. Indeed, alpha-synuclein (snca) is one of the major genes implicated in Parkinson's disease (PD). Howe... |