While a number of computational tools help design sgRNAs to target specific loci, the accurate prediction of whether Cas9 and the sgRNA of interest will bind specifically to the genome is still a challenge.
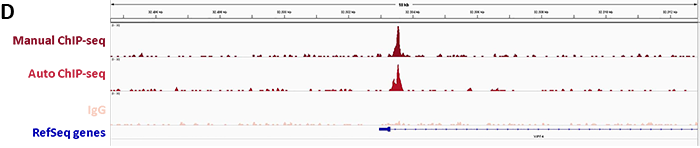
In order to ensure specific targeting of CRISPR/Cas9, the verification of the specific binding of the sgRNA at the locus of interest is required. The methodology of choice for such verification is chromatin immunoprecipitation followed either by real-time PCR (ChIP-qPCR) or sequencing (ChIP-seq) to analyze protein-DNA interactions.
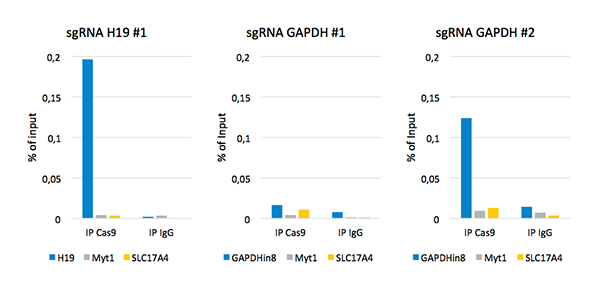
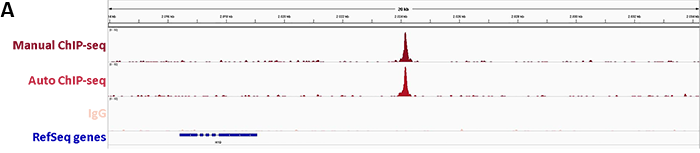
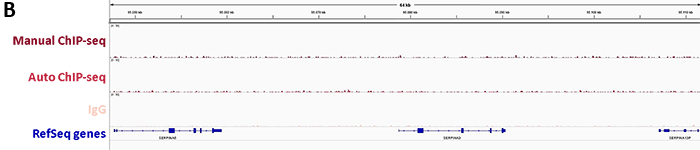
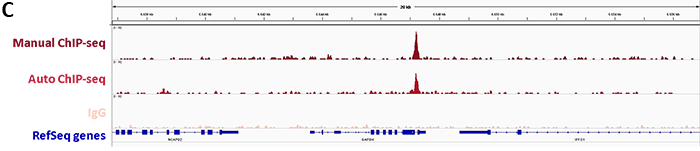
Diagenode’s iDeal ChIP-seq Kit for Transcription Factors provides a robust workflow to investigate dCas9 binding in the genome by specifically enriching dCas9 in the on-target region by ChIP followed by real-time PCR. In addition, sequencing analysis of the DNA immunoprecipitated with the Cas9 antibody illustrates potential genome-wide off-target effects. This information is essential to verify the correct binding of dCas9 fused to an effector.