The Megaruptor® was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA from 2 kb - 75 kb. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design.
TESTIMONIALAt the VIB Department of Molecular Genetics we strive to implement the best technologies in our workflows. Recent advances in long read sequencing using Oxford Nanopore technology have made the investigation of complex genomic variation in the human genome possible. A crucial step in the beginning of the library preparation is the shearing of genomic DNA, and this is where we implement the Megaruptor. The straightforward protocol is efficient, requires minimal sample handling and results in very reproducible fragment sizes. Another advantage is the consumable cost, which is lower than alternative technologies. For our applications we prefer shearing to lengths of 10 to 40kb, and for this the Megaruptor is the optimal solution.
Wouter de Coster and the Genomic Service Facility, VIB Department of Molecular Genetics, Antwerp, Belgium
TESTIMONIALThe Megaruptor® shows a more dense size distribution of sheared fragments, and a higher and more reproducible yield.
Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany
Morten Skage and Dr Ave Tooming-Klunderud, Norwegian Sequencing Centre, Oslo, NorwayThe Norwegian sequencing centre (NSC) is a national core facility fully equipped to handle a diverse set of sequencing projects. We see an increased interest in long read technologies, in which our PacBio Single Molecule sequencing service is important. PacBio, (or SMRT) sequencing, like any other single molecule system, starts with a critical library preparation step to generate long fragment libraries of 10-20kb length. Even longer libraries can be made, but common to all library types is to start with good quality DNA of high molecular weight. Physical shearing of the DNA, targeting the desired library length, is the first critical step in the procedure. NSC has experience with different methods to fragment DNA like nebulization, enzymatic, tagmentation, covaris AFA, g-tubes (also covaris), custom syringe/needle protocols and hydroshearing. The best fragmentation method should produce a sharp fragmentation pattern close to the target length as defined by the protocol settings, with as little “smear like” pattern of lower molecular length as possible. The final step of PacBio library preparation is to size select only the longest fragments (> 7kb) as this increases the overall success rate of only sequencing the longest library fragments. It is not uncommon to loose a lot of material during this step. However, if the initial fragmentation yields sharp fragmentation patterns with minimal smear, more of the total DNA is retained in the final library. This is perhaps most important on samples were DNA amounts are limited. The optimal fragmentation method has to produce similar fragmentation pattern each time, with little deviation with sample type and input amount. We have found the new Megaruptor from Diagenode to be one of the best instruments for routine shearing of DNA to lengths of 2-40kb. It is easy to use, with walk away shearing, and the instrument does all the washes before, in between samples and after shearing without user intervention. Disposable hydropores is beneficial for lowering the contamination risk. The fact that it is so easy to use make it a good choice for laboratories with multiple users and/or when training new staff.
We will use the Megaruptor® within our PacBio® Single Molecule Sequencing workflows, especially for the construction of very big insert (>20kb) libraries.
Andrea Patrignani, Anna Bratus and Ralph Schlapbach, Functional Genomics Center Zurich (ETHZ/UZH), Switzerland
| Improvements in Hydrodynamic Shearing Facilitates 20-50 kb True Mate Pair Library Construction POSTER The Megaruptor® has been used by several sequencing centers to produce high-quality small ins... | Download |
| Megaruptor® DNA Shearing System User Manual v2 MANUAL The Megaruptor® was designed to provide researchers with a simple, automated, and repro... | Download |
How to properly cite this product in your workDiagenode strongly recommends using this: Megaruptor® (Diagenode Cat# B06010001). Click here to copy to clipboard. Using our products in your publication? Let us know! |
Complete Genome Sequence of the Type Strain Pectobacterium punjabenseSS95, Isolated from a Potato Plant with Blackleg Symptoms. |
Sizing, stabilising, and cloning repeat-expansions for gene targeting constructs. |
Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. |
Nanopore Sequencing and Its Clinical Applications. |
Condition-dependent co-regulation of genomic clusters of virulence factors in the grapevine trunk pathogen Neofusicoccum parvum |
Complete genome of Staphylococcus aureus Tager 104 provides evidence of its relation to modern systemic hospital-acquired strains |
Complete Genome Sequences of Four Escherichia coli ST95 Isolates from Bloodstream Infections |
Diversification of bacterial genome content through distinct mechanisms over different timescales. |
Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance |
Microevolution of Burkholderia pseudomallei during an acute infection. |
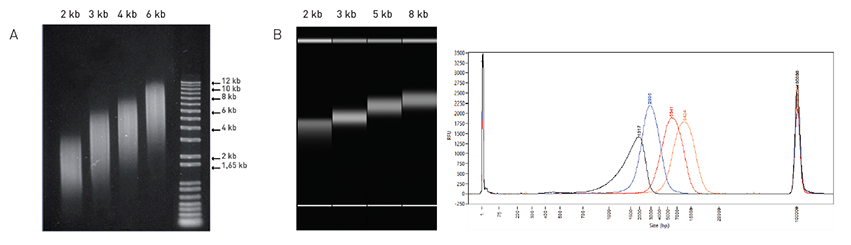
Notice (8): Undefined variable: solution_of_interest [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL --><div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"><?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>$viewFile = '/home/website-server/www/app/View/Products/view.ctp' $dataForView = array( 'language' => 'en', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'meta_title' => 'Megaruptor', 'product' => array( 'Product' => array( 'id' => '1824', 'antibody_id' => null, 'name' => 'Megaruptor<sup>®</sup>', 'description' => '<p><span>The Megaruptor</span><sup>®</sup><span> was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA from </span><strong>2 kb - 75 kb</strong><span>. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p>', 'label1' => 'Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores', 'info1' => '<h6><img src="https://www.diagenode.com/img/product/shearing_technologies/megaruptor-fig1.png" alt="" style="display: block; margin-left: auto; margin-right: auto;" width="851" height="245" /></h6> <h6 style="text-align: center;">Validation using two different DNA sources and two different methods of analysis. <strong>A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Fragment Analyzer profiles of human genomic DNA (25 ng/μl; 200 μl/sample) sheared at different software settings of 2, 3, 5 and 8 kb. (Standard Sensitivity Large Fragment Analysis Kit ; Advanced Analytical Technologies, Inc. was used for separation and fragment sizing).</h6>', 'label2' => '', 'info2' => '', 'label3' => '', 'info3' => '', 'format' => '1 unit', 'catalog_number' => 'B06010001', 'old_catalog_number' => '', 'sf_code' => null, 'type' => 'ACC', 'search_order' => '04-undefined', 'price_EUR' => 'on request', 'price_USD' => 'on request', 'price_GBP' => 'on request', 'price_JPY' => 'on request', 'price_CNY' => 'on request', 'price_AUD' => 'on request', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => true, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'megaruptor-1-unit', 'meta_title' => 'Megaruptor', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'modified' => '2019-02-20 12:43:01', 'created' => '2015-06-29 14:08:20', 'locale' => 'eng' ), 'Antibody' => array( 'host' => '*****', 'id' => null, 'name' => null, 'description' => null, 'clonality' => null, 'isotype' => null, 'lot' => null, 'concentration' => null, 'reactivity' => null, 'type' => null, 'purity' => null, 'classification' => null, 'application_table' => null, 'storage_conditions' => null, 'storage_buffer' => null, 'precautions' => null, 'uniprot_acc' => null, 'slug' => null, 'meta_keywords' => null, 'meta_description' => null, 'modified' => null, 'created' => null, 'select_label' => null ), 'Slave' => array(), 'Group' => array(), 'Related' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ) ), 'Application' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ) ), 'Category' => array(), 'Document' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ) ), 'Feature' => array(), 'Image' => array( (int) 0 => array( [maximum depth reached] ) ), 'Promotion' => array(), 'Protocol' => array(), 'Publication' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ), (int) 3 => array( [maximum depth reached] ), (int) 4 => array( [maximum depth reached] ), (int) 5 => array( [maximum depth reached] ), (int) 6 => array( [maximum depth reached] ), (int) 7 => array( [maximum depth reached] ), (int) 8 => array( [maximum depth reached] ), (int) 9 => array( [maximum depth reached] ) ), 'Testimonial' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ), (int) 3 => array( [maximum depth reached] ) ), 'Area' => array(), 'SafetySheet' => array() ) ) $language = 'en' $meta_keywords = '' $meta_description = 'Megaruptor' $meta_title = 'Megaruptor' $product = array( 'Product' => array( 'id' => '1824', 'antibody_id' => null, 'name' => 'Megaruptor<sup>®</sup>', 'description' => '<p><span>The Megaruptor</span><sup>®</sup><span> was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA from </span><strong>2 kb - 75 kb</strong><span>. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p>', 'label1' => 'Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores', 'info1' => '<h6><img src="https://www.diagenode.com/img/product/shearing_technologies/megaruptor-fig1.png" alt="" style="display: block; margin-left: auto; margin-right: auto;" width="851" height="245" /></h6> <h6 style="text-align: center;">Validation using two different DNA sources and two different methods of analysis. <strong>A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Fragment Analyzer profiles of human genomic DNA (25 ng/μl; 200 μl/sample) sheared at different software settings of 2, 3, 5 and 8 kb. (Standard Sensitivity Large Fragment Analysis Kit ; Advanced Analytical Technologies, Inc. was used for separation and fragment sizing).</h6>', 'label2' => '', 'info2' => '', 'label3' => '', 'info3' => '', 'format' => '1 unit', 'catalog_number' => 'B06010001', 'old_catalog_number' => '', 'sf_code' => null, 'type' => 'ACC', 'search_order' => '04-undefined', 'price_EUR' => 'on request', 'price_USD' => 'on request', 'price_GBP' => 'on request', 'price_JPY' => 'on request', 'price_CNY' => 'on request', 'price_AUD' => 'on request', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => true, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'megaruptor-1-unit', 'meta_title' => 'Megaruptor', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'modified' => '2019-02-20 12:43:01', 'created' => '2015-06-29 14:08:20', 'locale' => 'eng' ), 'Antibody' => array( 'host' => '*****', 'id' => null, 'name' => null, 'description' => null, 'clonality' => null, 'isotype' => null, 'lot' => null, 'concentration' => null, 'reactivity' => null, 'type' => null, 'purity' => null, 'classification' => null, 'application_table' => null, 'storage_conditions' => null, 'storage_buffer' => null, 'precautions' => null, 'uniprot_acc' => null, 'slug' => null, 'meta_keywords' => null, 'meta_description' => null, 'modified' => null, 'created' => null, 'select_label' => null ), 'Slave' => array(), 'Group' => array(), 'Related' => array( (int) 0 => array( 'id' => '2647', 'antibody_id' => null, 'name' => 'Hydro Tubes', 'description' => '<p><span>Hydro Tubes for Megaruptor</span><sup>®</sup><span> The Microcentrifuge Tubes (0.5 ml) have been thoroughly validated for use on the Megaruptor</span><sup>®</sup><span> (B06010001) and are strongly recommended for DNA shearing applications using this instrument.</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => 'Characteristics', 'info1' => '<ul> <li>Sterile</li> <li>Natural</li> <li>Autoclavable</li> <li>Freezable</li> </ul> <p></p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '50 pc', 'catalog_number' => 'C30010018', 'old_catalog_number' => '', 'sf_code' => 'C30010018-1860', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '65', 'price_USD' => '65', 'price_GBP' => '60', 'price_JPY' => '10180', 'price_CNY' => '', 'price_AUD' => '162', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydro-tubes-50-pc', 'meta_title' => 'Hydro Tubes', 'meta_keywords' => '', 'meta_description' => 'Hydro Tubes', 'modified' => '2020-04-07 05:19:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '2645', 'antibody_id' => null, 'name' => 'Hydropore - short', 'description' => '<p><span style="font-weight: 400;">Hydropore-shortは<strong>2 kb - 9 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010001', 'old_catalog_number' => '', 'sf_code' => 'E07010001-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '135', 'price_USD' => '160', 'price_GBP' => '125', 'price_JPY' => '21150', 'price_CNY' => '', 'price_AUD' => '400', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-short-10-pc', 'meta_title' => 'Hydropore - short', 'meta_keywords' => '', 'meta_description' => 'Hydropore - short', 'modified' => '2022-04-18 09:01:09', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '2646', 'antibody_id' => null, 'name' => 'Hydropore - long', 'description' => '<p><span style="font-weight: 400;">Hydropore-longは<strong>10 kb - 75 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010002', 'old_catalog_number' => '', 'sf_code' => 'E07010002-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '145', 'price_USD' => '170', 'price_GBP' => '130', 'price_JPY' => '22715', 'price_CNY' => '', 'price_AUD' => '425', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-long-10-pc', 'meta_title' => 'Hydropore - long', 'meta_keywords' => '', 'meta_description' => 'Hydropore - long', 'modified' => '2022-04-18 09:02:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ) ), 'Application' => array( (int) 0 => array( 'id' => '13', 'position' => '10', 'parent_id' => '3', 'name' => 'DNA/RNA shearing', 'description' => '<div class="row"> <div class="small-12 medium-12 large-12 columns">In recent years, advances in Next-Generation Sequencing (NGS) have revolutionized genomics and biology. This growth has fueled demands on upstream techniques for optimal sample preparation and genomic library construction. One of the most critical aspects of optimal library preparation is the quality of the DNA to be sequenced. The DNA must first be effectively and consistently sheared into the appropriate fragment size (depending on the sequencing platform) to enable sensitive and reliable NGS results. The <strong>Bioruptor</strong><sup>®</sup> <strong>Pico</strong> and the <strong>Megaruptor</strong><sup>®</sup> provide superior sample yields, fragment size, and consistency, which are essential for Next-Generation Sequencing workflows. Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor<sup>®</sup></a>.</div> </div> <p></p> <div class="row"> <div class="small-7 medium-7 large-7 columns text-center"><img src="https://www.diagenode.com/img/applications/true-flexibility-with-br-ngs.jpg" /></div> <div class="small-5 medium-5 large-5 columns"><small><strong>Programmable DNA size distribution and high reproducibility with Bioruptor<sup>®</sup> Pico using 0.65 (panel A) or 0.1 ml (panel B) microtubes</strong>. <b>Panel A:</b> 200 bp after 13 cycles (13 sec ON/OFF) using 100 µl volume. Average size: 204; CV%:1.89%). <b>Panel B:</b> 200 bp after 20 cycles (30 sec ON/OFF) using 10 µl volume. (Average size: 215 bp; CV%: 6.6%). <b>Panel A & B:</b> peak electropherogram view. <b>Panel C & D:</b> virtual gel view.</small></div> </div> <p><br /><br /></p> <div class="row"> <div class="small-10 medium-10 large-10 columns text-center end small-offset-1"><img src="https://www.diagenode.com/img/applications/megaruptor-short-frag.jpg" /></div> <div class="small-12 medium-12 large-12 columns"><small><strong> Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores Validation using two different DNA sources and two different methods of analysis. A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Bioanalyzer profiles of human genomic DNA (20 ng/μl; 150 μl/sample) sheared at different software settings of 2 and 5 kb. Three independent experiments were run for each setting. (Agilent DNA 12000 kit was used for separation and fragment sizing).</small></div> </div> <p><br /><br /></p> <div class="row"> <div class="small-4 medium-4 large-4 columns text-center"><img src="https://www.diagenode.com/img/applications/megaruptor-long-frag.jpg" /></div> <div class="small-8 medium-8 large-8 columns"><small><strong> Demonstrated shearing to fragment sizes between 15 kb and 75 kb with Megaruptor® using long fragment size Hydropores. </strong>Image shows DNA size distribution of human genomic DNA sheared with long fragment Hydropores. DNA was analyzed by pulsed field gel electrophoresis (PFGE) in 1% agarose gel and a mean size of smears was estimated using Image Lab 4.1 software.<br /> * indicates unsheared DNA </small></div> </div>', 'in_footer' => false, 'in_menu' => true, 'online' => true, 'tabular' => false, 'slug' => 'dna-rna-shearing', 'meta_keywords' => 'DNA/RNA shearing,Bioruptor® Pico,Megaruptor®,Next-Generation Sequencing ', 'meta_description' => 'Bioruptor® Pico and the Megaruptor® provide superior sample yields, fragment size, and consistency, which are essential for Next-Generation Sequencing workflows.', 'meta_title' => 'DNA shearing & RNA shearing for Next-Generation Sequencing (NGS) | Diagenode', 'modified' => '2017-12-08 14:44:11', 'created' => '2014-10-29 12:45:41', 'ProductsApplication' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '3', 'position' => '10', 'parent_id' => null, 'name' => '次世代シーケンシング', 'description' => '<div class="row"> <div class="small-12 medium-12 large-12 columns"> <h2 style="font-size: 22px;">DNA断片化、ライブラリー調製、自動化:NGSのワンストップショップ</h2> <table class="small-12 medium-12 large-12 columns"> <tbody> <tr> <th class="small-12 medium-12 large-12 columns"> <h4>1. 断片化装置を選択してください:150 bp〜75 kbの範囲でDNAを断片化します。</h4> </th> </tr> <tr style="background-color: #ffffff;"> <td class="small-12 medium-12 large-12 columns"></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns"><a href="../p/bioruptor-pico-sonication-device"><img src="https://www.diagenode.com/img/product/shearing_technologies/bioruptor_pico.jpg" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/megaruptor2-1-unit"><img src="https://www.diagenode.com/img/product/shearing_technologies/B06010001_megaruptor2.jpg" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/bioruptor-one-sonication-device"><img src="https://www.diagenode.com/img/product/shearing_technologies/br-one-profil.png" /></a></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns">5μlまで断片化:150 bp〜2 kb<br />NGS DNAライブラリー調製およびFFPE核酸抽出に最適で、</td> <td class="small-4 medium-4 large-4 columns">2 kb〜75 kbの範囲をできます。<br />メイトペアライブラリー調製および長いフラグメントDNAシーケンシングに最適で、この軽量デスクトップデバイスで</td> <td class="small-4 medium-4 large-4 columns">20または50μlの断片化が可能です。</td> </tr> </tbody> </table> <table class="small-12 medium-12 large-12 columns"> <tbody> <tr> <th class="small-8 medium-8 large-8 columns"> <h4>2. 最適化されたライブラリー調整キットを選択してください。</h4> </th> <th class="small-4 medium-4 large-4 columns"> <h4>3. ライブラリー前処理自動化を選択して、比類のないデータ再現性を実感</h4> </th> </tr> <tr style="background-color: #ffffff;"> <td class="small-12 medium-12 large-12 columns"></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns"><a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns"><img src="https://www.diagenode.com/img/product/kits/microPlex_library_preparation.png" style="display: block; margin-left: auto; margin-right: auto;" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/ideal-library-preparation-kit-x24-incl-index-primer-set-1-24-rxns"><img src="https://www.diagenode.com/img/product/kits/box_kit.jpg" style="display: block; margin-left: auto; margin-right: auto;" height="173" width="250" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/sx-8g-ip-star-compact-automated-system-1-unit"><img src="https://www.diagenode.com/img/product/automation/B03000002%20_ipstar_compact.png" style="display: block; margin-left: auto; margin-right: auto;" /></a></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">50pgの低入力:MicroPlex Library Preparation Kit</td> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">5ng以上:iDeal Library Preparation Kit</td> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">Achieve great NGS data easily</td> </tr> </tbody> </table> </div> </div> <blockquote> <div class="row"> <div class="small-12 medium-12 large-12 columns"><span class="label" style="margin-bottom: 16px; margin-left: -22px; font-size: 15px;">DiagenodeがNGS研究にぴったりなプロバイダーである理由</span> <p>Diagenodeは15年以上もエピジェネティクス研究に専念、専門としています。 ChIP研究クロマチン用のユニークな断片化システムの開発から始まり、 専門知識を活かし、5μlのせん断体積まで可能で、NGS DNAライブラリーの調製に最適な最先端DNA断片化装置の開発にたどり着きました。 我々は以来、ChIP-seq、Methyl-seq、NGSライブラリー調製用キットを研究開発し、業界をリードする免疫沈降研究と同様に、ライブラリー調製を自動化および完結させる独自の自動化システムを開発にも成功しました。</p> <ul> <li>信頼されるせん断装置</li> <li>様々なインプットからのライブラリ作成キット</li> <li>独自の自動化デバイス</li> </ul> </div> </div> </blockquote> <div class="row"> <div class="small-12 columns"> <ul class="accordion" data-accordion=""> <li class="accordion-navigation"><a href="#panel1a">次世代シーケンシングへの理解とその専門知識</a> <div id="panel1a" class="content"> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p><strong>次世代シーケンシング (NGS)</strong> )は、著しいスケールとハイスループットでシーケンシングを行い、1日に数十億もの塩基生成を可能にします。 NGSのハイスループットは迅速でありながら正確で、再現性のあるデータセットを実現し、さらにシーケンシング費用を削減します。 NGSは、ゲノムシーケンシング、ゲノム再シーケンシング、デノボシーケンシング、トランスクリプトームシーケンシング、その他にDNA-タンパク質相互作用の検出やエピゲノムなどを示します。 指数関数的に増加するシーケンシングデータの需要は、計算分析の障害や解釈、データストレージなどの課題を解決します。</p> <p>アプリケーションおよび出発物質に応じて、数百万から数十億の鋳型DNA分子を大規模に並行してシーケンシングすることが可能です。その為に、異なる化学物質を使用するいくつかの市販のNGSプラットフォームを利用することができます。 NGSプラットフォームの種類によっては、事前準備とライブラリー作成が必要です。</p> <p>NGSにとっても、特にデータ処理と分析に関した大きな課題はあります。第3世代技術はゲノミクス研究にさらに革命を起こすであろうと大きく期待されています。</p> </div> </div> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <p><strong>NGS アプリケーション</strong></p> <ul> <li>全ゲノム配列決定</li> <li>デノボシーケンシング</li> <li>標的配列</li> <li>Exomeシーケンシング</li> <li>トランスクリプトーム配列決定</li> <li>ゲノム配列決定</li> <li>ミトコンドリア配列決定</li> <li>DNA-タンパク質相互作用(ChIP-seq</li> <li>バリアント検出</li> <li>ゲノム仕上げ</li> </ul> </div> <div class="small-6 medium-6 large-6 columns"> <p><strong>研究分野におけるNGS:</strong></p> <ul> <li>腫瘍学</li> <li>リプロダクティブ・ヘルス</li> <li>法医学ゲノミクス</li> <li>アグリゲノミックス</li> <li>複雑な病気</li> <li>微生物ゲノミクス</li> <li>食品・環境ゲノミクス</li> <li>創薬ゲノミクス - パーソナライズド・メディカル</li> </ul> </div> <div class="small-12 medium-12 large-12 columns"> <p><strong>NGSの用語</strong></p> <dl> <dt>リード(読み取り)</dt> <dd>この装置から得られた連続した単一のストレッチ</dd> <dt>断片リード</dt> <dd>フラグメントライブラリからの読み込み。 シーケンシングプラットフォームに応じて、読み取りは通常約100〜300bp。</dd> <dt>断片ペアエンドリード</dt> <dd>断片ライブラリーからDNA断片の各末端2つの読み取り。</dd> <dt>メイトペアリード</dt> <dd>大きなDNA断片(通常は予め定義されたサイズ範囲)の各末端から2つの読み取り。</dd> <dt>カバレッジ(例)</dt> <dd>30×適用範囲とは、参照ゲノム中の各塩基対が平均30回の読み取りを示す。</dd> </dl> </div> </div> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <h2>NGSプラットフォーム</h2> <h3><a href="http://www.illumina.com" target="_blank">イルミナ</a></h3> <p>イルミナは、クローン的に増幅された鋳型DNA(クラスター)上に位置する、蛍光標識された可逆的鎖ターミネーターヌクレオチドを用いた配列別合成技術を使用。 DNAクラスターは、ガラスフローセルの表面上に固定化され、 ワークフローは、4つのヌクレオチド(それぞれ異なる蛍光色素で標識された)の組み込み、4色イメージング、色素や末端基の切断、取り込み、イメージングなどを繰り返します。フローセルは大規模な並列配列決定を受ける。 この方法により、単一蛍光標識されたヌクレオチドの制御添加によるモノヌクレオチドのエラーを回避する可能性があります。 読み取りの長さは、通常約100〜150 bpです。</p> <h3><a href="http://www.lifetechnologies.com" target="_blank">イオン トレント</a></h3> <p>イオントレントは、半導体技術チップを用いて、合成中にヌクレオチドを取り込む際に放出されたプロトンを検出します。 これは、イオン球粒子と呼ばれるビーズの表面にエマルションPCR(emPCR)を使用し、リンクされた特定のアダプターを用いてDNA断片を増幅します。 各ビーズは1種類のDNA断片で覆われていて、異なるDNA断片を有するビーズは次いで、チップの陽子感知ウェル内に配置されます。 チップには一度に4つのヌクレオチドのうちの1つが浸水し、このプロセスは異なるヌクレオチドで15秒ごとに繰り返されます。 配列決定の間に4つの塩基の各々が1つずつ導入されます、組み込みの場合はプロトンが放出され、電圧信号が取り込みに比例して検出されます。.</p> <h3><a href="http://www.pacificbiosciences.com" target="_blank">パシフィック バイオサイエンス</a></h3> <p>パシフィックバイオサイエンスでは、20kbを超える塩基対の読み取りも、単一分子リアルタイム(SMRT)シーケンシングによる構造および細胞タイプの変化を観察することができます。 このプラットフォームでは、超長鎖二本鎖DNA(dsDNA)断片が、Megaruptor(登録商標)のようなDiagenode装置を用いたランダムシアリングまたは目的の標的領域の増幅によって生成されます。 SMRTbellライブラリーは、ユニバーサルヘアピンアダプターをDNA断片の各末端に連結することによって生成します。 サイズ選択条件による洗浄ステップの後、配列決定プライマーをSMRTbellテンプレートにアニーリングし、鋳型DNAに結合したDNAポリメラーゼを含む配列決定を、蛍光標識ヌクレオチドの存在下で開始。 各塩基が取り込まれると、異なる蛍光のパルスをリアルタイムで検出します。</p> <h3><a href="https://nanoporetech.com" target="_blank">オックスフォード ナノポア</a></h3> <p>Oxford Nanoporeは、単一のDNA分子配列決定に基づく技術を開発します。その技術により生物学的分子、すなわちDNAが一群の電気抵抗性高分子膜として位置するナノスケールの孔(ナノ細孔)またはその近くを通過し、イオン電流が変化します。 この変化に関する情報は、例えば4つのヌクレオチド(AまたはG r CまたはT)ならびに修飾されたヌクレオチドすべてを区別することによって分子情報に訳されます。 シーケンシングミニオンデバイスのフローセルは、数百個のナノポアチャネルのセンサアレイを含みます。 DNAサンプルは、Diagenode社のMegaruptor(登録商標)を用いてランダムシアリングによって生成され得る超長鎖DNAフラグメントが必要です。</p> <h3><a href="http://www.lifetechnologies.com/be/en/home/life-science/sequencing/next-generation-sequencing/solid-next-generation-sequencing.html" target="_blank">SOLiD</a></h3> <p>SOLiDは、ユニークな化学作用により、何千という個々のDNA分子の同時配列決定を可能にします。 それは、アダプター対ライブラリーのフラグメントが適切で、せん断されたゲノムDNAへのアダプターのライゲーションによるライブラリー作製から始まります。 次のステップでは、エマルジョンPCR(emPCR)を実施して、ビーズの表面上の個々の鋳型DNA分子をクローン的に増幅。 emPCRでは、個々の鋳型DNAをPCR試薬と混合し、水中油型エマルジョン内の疎水性シェルで囲まれた水性液滴内のプライマーコートビーズを、配列決定のためにロードするスライドガラスの表面にランダムに付着。 この技術は、シークエンシングプライマーへのライゲーションで競合する4つの蛍光標識されたジ塩基プローブのセットを使用します。</p> <h3><a href="http://454.com/products/technology.asp" target="_blank">454</a></h3> <p>454は、大規模並列パイロシーケンシングを利用しています。 始めに全ゲノムDNAまたは標的遺伝子断片の300〜800bp断片のライブラリー調製します。 次に、DNAフラグメントへのアダプターの付着および単一のDNA鎖の分離。 その後アダプターに連結されたDNAフラグメントをエマルジョンベースのクローン増幅(emPCR)で処理し、DNAライブラリーフラグメントをミクロンサイズのビーズ上に配置します。 各DNA結合ビーズを光ファイバーチップ上のウェルに入れ、器具に挿入します。 4つのDNAヌクレオチドは、配列決定操作中に固定された順序で連続して加えられ、並行して配列決定されます。</p> </div> </div> </div> </li> </ul> </div> </div>', 'in_footer' => true, 'in_menu' => true, 'online' => true, 'tabular' => true, 'slug' => 'next-generation-sequencing', 'meta_keywords' => 'Next-generation sequencing,NGS,Whole genome sequencing,NGS platforms,DNA/RNA shearing', 'meta_description' => 'Diagenode offers kits and DNA/RNA shearing technology prior to next-generation sequencing, many Next-generation sequencing platforms require preparation of the DNA.', 'meta_title' => 'Next-generation sequencing (NGS) Applications and Methods | Diagenode', 'modified' => '2018-07-26 05:24:29', 'created' => '2015-04-01 22:47:04', 'ProductsApplication' => array( [maximum depth reached] ) ) ), 'Category' => array(), 'Document' => array( (int) 0 => array( 'id' => '14', 'name' => 'Improvements in Hydrodynamic Shearing Facilitates 20-50 kb True Mate Pair Library Construction', 'description' => '<p><span>The Megaruptor<sup>®</sup> has been used by several sequencing centers to produce high-quality small insert fragment libraries for Illumina and PacBio platforms. David Mead and colleagues from Lucigen<sup>®</sup> have used the Megaruptor<sup>®</sup> to prepare 20-50 kb inserts for a new mate-pair library technology that is >70% efficient. </span></p>', 'image_id' => null, 'type' => 'Poster', 'url' => 'files/posters/Mead_et_al_Lucigen_Diagenode_Poster.pdf', 'slug' => 'mead-et-al-lucigen-diagenode-poster', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-09-29 13:42:27', 'created' => '2015-07-03 16:05:15', 'ProductsDocument' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '783', 'name' => 'Megaruptor® DNA Shearing System User Manual v2', 'description' => '<div class="page" title="Page 4"> <div class="layoutArea"> <div class="column"> <p><span>The Megaruptor</span><span>® </span><span>was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA in the range of 2 kb - 75 kb. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p> </div> </div> </div>', 'image_id' => null, 'type' => 'Manual', 'url' => 'files/products/shearing_technology/megaruptor/Megaruptor_manual.pdf', 'slug' => 'megaruptor-manual', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-07-23 17:29:32', 'created' => '2015-07-07 11:47:45', 'ProductsDocument' => array( [maximum depth reached] ) ) ), 'Feature' => array(), 'Image' => array( (int) 0 => array( 'id' => '139', 'name' => 'product/shearing_technologies/B06010001_megaruptor.jpg', 'alt' => 'megaruptor', 'modified' => '2015-07-23 17:39:35', 'created' => '2015-06-10 17:28:55', 'ProductsImage' => array( [maximum depth reached] ) ) ), 'Promotion' => array(), 'Protocol' => array(), 'Publication' => array( (int) 0 => array( 'id' => '4018', 'name' => 'Complete Genome Sequence of the Type Strain Pectobacterium punjabenseSS95, Isolated from a Potato Plant with Blackleg Symptoms.', 'authors' => 'Sarfraz, S and Oulghazi, S and Cigna, J and Sahi, ST and Riaz, K andTufail, MR and Fayyaz, A and Naveed, K and Hameed, A and Lopez-Roques, Cand Vandecasteele, C and Faure, D', 'description' => '<p>Pectobacterium punjabense is a newly described species causing blackleg disease in potato plants. Therefore, by the combination of long (Oxford Nanopore Technologies, MinION) and short (Illumina MiSeq) reads, we sequenced the complete genome of P. punjabense SS95T, which contains a circular chromosome of 4.793 Mb with a GC content of 50.7\%.</p>', 'date' => '2020-08-06', 'pmid' => 'http://www.pubmed.gov/32763925', 'doi' => '10.1128/MRA.00420-20.', 'modified' => '2020-12-16 17:37:51', 'created' => '2020-10-12 14:54:59', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '4001', 'name' => 'Sizing, stabilising, and cloning repeat-expansions for gene targeting constructs.', 'authors' => 'Nair RR, Tibbit C, Thompson D, McLeod R, Nakhuda A, Simon MM, Baloh RH, Fisher EMC, Isaacs AM, Cunningham TJ', 'description' => '<p>Aberrant microsatellite repeat-expansions at specific loci within the human genome cause several distinct, heritable, and predominantly neurological, disorders. Creating models for these diseases poses a challenge, due to the instability of such repeats in bacterial vectors, especially with large repeat expansions. Designing constructs for more precise genome engineering projects, such as engineering knock-in mice, proves a greater challenge still, since these unstable repeats require numerous cloning steps in order to introduce homology arms or selection cassettes. Here, we report our efforts to clone a large hexanucleotide repeat in the C9orf72 gene, originating from within a BAC construct, derived from a C9orf72-ALS patient. We provide detailed methods for efficient repeat sizing and growth conditions in bacteria to facilitate repeat retention during growth and sub-culturing. We report that sub-cloning into a linear vector dramatically improves stability, but is dependent on the relative orientation of DNA replication through the repeat, consistent with previous studies. We envisage the findings presented here provide a relatively straightforward route to maintaining large-range microsatellite repeat-expansions, for efficient cloning into vectors.</p>', 'date' => '2020-07-25', 'pmid' => 'http://www.pubmed.gov/32721467', 'doi' => '10.1016/j.ymeth.2020.07.007', 'modified' => '2020-09-01 14:39:41', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '3980', 'name' => 'Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants.', 'authors' => 'Xu Z, Pu X, Gao R, Demurtas OC, Fleck SJ, Richter M, He C, Ji A, Sun W, Kong J, Hu K, Ren F, Song J, Wang Z, Gao T, Xiong C, Yu H, Xin T, Albert VA, Giuliano G, Chen S, Song J', 'description' => '<p>BACKGROUND: Plants have evolved a panoply of specialized metabolites that increase their environmental fitness. Two examples are caffeine, a purine psychotropic alkaloid, and crocins, a group of glycosylated apocarotenoid pigments. Both classes of compounds are found in a handful of distantly related plant genera (Coffea, Camellia, Paullinia, and Ilex for caffeine; Crocus, Buddleja, and Gardenia for crocins) wherein they presumably evolved through convergent evolution. The closely related Coffea and Gardenia genera belong to the Rubiaceae family and synthesize, respectively, caffeine and crocins in their fruits. RESULTS: Here, we report a chromosomal-level genome assembly of Gardenia jasminoides, a crocin-producing species, obtained using Oxford Nanopore sequencing and Hi-C technology. Through genomic and functional assays, we completely deciphered for the first time in any plant the dedicated pathway of crocin biosynthesis. Through comparative analyses with Coffea canephora and other eudicot genomes, we show that Coffea caffeine synthases and the first dedicated gene in the Gardenia crocin pathway, GjCCD4a, evolved through recent tandem gene duplications in the two different genera, respectively. In contrast, genes encoding later steps of the Gardenia crocin pathway, ALDH and UGT, evolved through more ancient gene duplications and were presumably recruited into the crocin biosynthetic pathway only after the evolution of the GjCCD4a gene. CONCLUSIONS: This study shows duplication-based divergent evolution within the coffee family (Rubiaceae) of two characteristic secondary metabolic pathways, caffeine and crocin biosynthesis, from a common ancestor that possessed neither complete pathway. These findings provide significant insights on the role of tandem duplications in the evolution of plant specialized metabolism.</p>', 'date' => '2020-06-18', 'pmid' => 'http://www.pubmed.gov/32552824', 'doi' => '10.1186/s12915-020-00795-3', 'modified' => '2020-09-01 15:27:14', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 3 => array( 'id' => '4002', 'name' => 'Nanopore Sequencing and Its Clinical Applications.', 'authors' => 'Sun X, Song L, Yang W, Zhang L, Liu M, Li X, Tian G, Wang W', 'description' => '<p>Nanopore sequencing is a method for determining the order and modifications of DNA/RNA nucleotides by detecting the electric current variations when DNA/RNA oligonucleotides pass through the nanometer-sized hole (nanopore). Nanopore-based DNA analysis techniques have been commercialized by Oxford Nanopore Technologies, NabSys, and Sequenom, and widely used in scientific researches recently including human genomics, cancer, metagenomics, plant sciences, etc., moreover, it also has potential applications in the field of healthcare due to its fast turn-around time, portable and real-time data analysis. Those features make it a promising technology for the point-of-care testing (POCT) and its potential clinical applications are briefly discussed in this chapter.</p>', 'date' => '2020-01-01', 'pmid' => 'http://www.pubmed.gov/32710311', 'doi' => '10.1007/978-1-0716-0904-0_2,', 'modified' => '2020-09-01 14:39:13', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 4 => array( 'id' => '3045', 'name' => 'Condition-dependent co-regulation of genomic clusters of virulence factors in the grapevine trunk pathogen Neofusicoccum parvum', 'authors' => 'Massonnet M. et al.', 'description' => '<p>The ascomycete Neofusicoccum parvum, one of the causal agents of Botryosphaeria dieback, is a destructive wood-infecting fungus and a serious threat to grape production worldwide. The capability of colonizing woody tissue combined with the secretion of phytotoxic compounds is thought to underlie its pathogenicity and virulence. Here, we describe the repertoire of virulence factors and their transcriptional dynamics as the fungus feeds on different substrates and colonizes the woody stem. We assembled and annotated a highly contiguous genome using single molecule real-time DNA sequencing. Transcriptome profiling by RNA-sequencing determined the genome-wide patterns of expression of virulence factors both in vitro (potato dextrose agar or medium amended with grape wood as substrate) and in planta. Pairwise statistical testing of differential expression followed by co-expression network analysis revealed that physically clustered genes coding for putative virulence functions were induced depending on substrate or stage of plant infection. Co-expressed gene clusters were significantly enriched not only in genes associated with secondary metabolism, but also with cell wall degradation, suggesting that dynamic co-regulation of transcriptional networks contribute to multiple aspects of N. parvum virulence. In most of the co-expressed clusters, all genes shared at least a common motif in their promoter region indicative of co-regulation by the same transcription factor. Co-expression analysis also identified chromatin regulators with correlated expression with inducible clusters of virulence factors, suggesting a complex, multi-layered regulation of the virulence repertoire of N. parvum.</p>', 'date' => '2016-09-08', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27608421', 'doi' => '', 'modified' => '2016-10-10 10:53:20', 'created' => '2016-10-10 10:53:20', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 5 => array( 'id' => '2839', 'name' => 'Complete genome of Staphylococcus aureus Tager 104 provides evidence of its relation to modern systemic hospital-acquired strains', 'authors' => 'Richard W. DavisIV, Andrew D. Brannen, Mohammad J. Hossain, Scott Monsma, Paul E. Bock, Matthias Nahrendorf, David Mead, Michael Lodes, Mark R. Liles and Peter Panizzi', 'description' => '<section xmlns="" xmlns:meta="http://www.springer.com/app/meta" class="Abstract" id="Abs1" lang="en"> <div class="js-CollapseSection"> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec1"> <h5 class="Heading">Background</h5> <p id="Par1" class="Para"><em xmlns="" class="EmphasisTypeItalic">Staphylococcus aureus</em> (<em xmlns="" class="EmphasisTypeItalic">S. aureus</em>) infections range in severity due to expression of certain virulence factors encoded on mobile genetic elements (MGE). As such, characterization of these MGE, as well as single nucleotide polymorphisms, is of high clinical and microbiological importance. To understand the evolution of these dangerous pathogens, it is paramount to define reference strains that may predate MGE acquisition. One such candidate is <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> Tager 104, a previously uncharacterized strain isolated from a patient with impetigo in 1947.</p> </div> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec2"> <h5 class="Heading">Results</h5> <p id="Par2" class="Para">We show here that <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> Tager 104 can survive in the bloodstream and infect naïve organs. We also demonstrate a procedure to construct and validate the assembly of <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> genomes, using Tager 104 as a proof-of-concept. In so doing, we bridged confounding gap regions that limited our initial attempts to close this 2.82 Mb genome, through integration of data from Illumina Nextera paired-end, PacBio RS, and Lucigen NxSeq mate-pair libraries. Furthermore, we provide independent confirmation of our segmental arrangement of the Tager 104 genome by the sole use of Lucigen NxSeq libraries filled by paired-end MiSeq reads and alignment with SPAdes software. Genomic analysis of Tager 104 revealed limited MGE, and a νSaβ island configuration that is reminiscent of other hospital acquired <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> genomes.</p> </div> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec3"> <h5 class="Heading">Conclusions</h5> <p id="Par3" class="Para">Tager 104 represents an early-branching ancestor of certain hospital-acquired strains. Combined with its earlier isolation date and limited content of MGE, Tager 104 can serve as a viable reference for future comparative genome studies.</p> </div> </div> </section>', 'date' => '2016-03-03', 'pmid' => 'http://pubmed.gov/26940863', 'doi' => '10.1186/s12864-016-2433-8', 'modified' => '2016-03-09 10:13:09', 'created' => '2016-03-09 10:13:09', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 6 => array( 'id' => '2953', 'name' => 'Complete Genome Sequences of Four Escherichia coli ST95 Isolates from Bloodstream Infections', 'authors' => 'Craig M. Stephens, Jeffrey M. Skerker, Manraj S. Sekhon, Adam P. Arkin, Lee W. Rileyd', 'description' => '<p>Finished genome sequences are presented for four Escherichia coli strains isolated from bloodstream infections at San Francisco General Hospital. These strains provide reference sequences for four major fimH-identified sublineages within the multilocus sequence type (MLST) ST95 group, and provide insights into pathogenicity and differential antimicrobial susceptibility within this group.</p>', 'date' => '2015-11-05', 'pmid' => 'http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4645194/', 'doi' => '10.1128/genomeA.01241-15', 'modified' => '2016-06-13 15:50:25', 'created' => '2016-06-13 15:50:25', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 7 => array( 'id' => '2372', 'name' => 'Diversification of bacterial genome content through distinct mechanisms over different timescales.', 'authors' => 'Croucher NJ, Coupland PG, Stevenson AE, Callendrello A, Bentley SD, Hanage WP', 'description' => 'Bacterial populations often consist of multiple co-circulating lineages. Determining how such population structures arise requires understanding what drives bacterial diversification. Using 616 systematically sampled genomes, we show that Streptococcus pneumoniae lineages are typically characterized by combinations of infrequently transferred stable genomic islands: those moving primarily through transformation, along with integrative and conjugative elements and phage-related chromosomal islands. The only lineage containing extensive unique sequence corresponds to a set of atypical unencapsulated isolates that may represent a distinct species. However, prophage content is highly variable even within lineages, suggesting frequent horizontal transmission that would necessitate rapidly diversifying anti-phage mechanisms to prevent these viruses sweeping through populations. Correspondingly, two loci encoding Type I restriction-modification systems able to change their specificity over short timescales through intragenomic recombination are ubiquitous across the collection. Hence short-term pneumococcal variation is characterized by movement of phage and intragenomic rearrangements, with the slower transfer of stable loci distinguishing lineages.', 'date' => '2014-11-19', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25407023', 'doi' => '', 'modified' => '2015-07-24 15:39:04', 'created' => '2015-07-24 15:39:04', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 8 => array( 'id' => '2838', 'name' => 'Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance', 'authors' => 'Mark R Davies, Matthew T Holden, Paul Coupland, Jonathan H K Chen, Carola Venturini, Timothy C Barnett, Nouri L Ben Zakour, Herman Tse, Gordon Dougan, Kwok-Yung Yuen & Mark J Walker', 'description' => '<p><span>A scarlet fever outbreak began in mainland China and Hong Kong in 2011 (refs. </span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref1" title="Chen, M. et al. Outbreak of scarlet fever associated with emm12 type group A Streptococcus in 2011 in Shanghai, China. Pediatr. Infect. Dis. J. 31, e158-e162 (2012)." id="ref-link-1">1</a><span>–</span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref6" title="Yang, P. et al. Characteristics of group A Streptococcus strains circulating during scarlet fever epidemic, Beijing, China, 2011. Emerg. Infect. Dis. 19, 909-915 (2013)." id="ref-link-2">6</a><span>). Macrolide- and tetracycline-resistant </span><i>Streptococcus pyogenes emm</i><span>12 isolates represent the majority of clinical cases. Recently, we identified two mobile genetic elements that were closely associated with </span><i>emm</i><span>12 outbreak isolates: the integrative and conjugative element ICE-</span><i>emm</i><span>12, encoding genes for tetracycline and macrolide resistance, and prophage </span><span class="mb">Φ</span><span>HKU.vir, encoding the superantigens SSA and SpeC, as well as the DNase Spd1 (ref. </span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref4" title="Tse, H. et al. Molecular characterization of the 2011 Hong Kong scarlet fever outbreak. J. Infect. Dis. 206, 341-351 (2012)." id="ref-link-3">4</a><span>). Here we sequenced the genomes of 141 </span><i>emm</i><span>12 isolates, including 132 isolated in Hong Kong between 2005 and 2011. We found that the introduction of several ICE-</span><i>emm</i><span>12 variants, </span><span class="mb">Φ</span><span>HKU.vir and a new prophage, </span><span class="mb">Φ</span><span>HKU.ssa, occurred in three distinct </span><i>emm</i><span>12 lineages late in the twentieth century. Acquisition of </span><i>ssa</i><span> and transposable elements encoding multidrug resistance genes triggered the expansion of scarlet fever–associated </span><i>emm</i><span>12 lineages in Hong Kong. The occurrence of multidrug-resistant </span><i>ssa</i><span>-harboring scarlet fever strains should prompt heightened surveillance within China and abroad for the dissemination of these mobile genetic elements.</span></p>', 'date' => '2014-11-17', 'pmid' => 'http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html', 'doi' => '10.1038/ng.3147', 'modified' => '2016-03-08 14:33:49', 'created' => '2016-03-08 14:33:21', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 9 => array( 'id' => '2262', 'name' => 'Microevolution of Burkholderia pseudomallei during an acute infection.', 'authors' => 'Limmathurotsakul D, Holden MT, Coupland P, Price EP, Chantratita N, Wuthiekanun V, Amornchai P, Parkhill J, Peacock SJ', 'description' => 'We used whole-genome sequencing to evaluate 69 independent colonies of Burkholderia pseudomallei isolated from seven body sites of a patient with acute disseminated melioidosis. Fourteen closely related genotypes were found, providing evidence for the rapid in vivo diversification of B. pseudomallei after inoculation and systemic spread.', 'date' => '2014-09-01', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24966357', 'doi' => '', 'modified' => '2015-07-24 15:39:03', 'created' => '2015-07-24 15:39:03', 'ProductsPublication' => array( [maximum depth reached] ) ) ), 'Testimonial' => array( (int) 0 => array( 'id' => '57', 'name' => 'VIB - Megaruptor', 'description' => '<p>At the VIB Department of Molecular Genetics we strive to implement the best technologies in our workflows. Recent advances in long read sequencing using Oxford Nanopore technology have made the investigation of complex genomic variation in the human genome possible. A crucial step in the beginning of the library preparation is the shearing of genomic DNA, and this is where we implement the Megaruptor. The straightforward protocol is efficient, requires minimal sample handling and results in very reproducible fragment sizes. Another advantage is the consumable cost, which is lower than alternative technologies. For our applications we prefer shearing to lengths of 10 to 40kb, and for this the Megaruptor is the optimal solution.</p>', 'author' => 'Wouter de Coster and the Genomic Service Facility, VIB Department of Molecular Genetics, Antwerp, Belgium', 'featured' => true, 'slug' => 'vib-antwerp', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:35:57', 'created' => '2016-07-06 10:35:57', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '42', 'name' => 'Norwegian Sequencing Center - Megaruptor', 'description' => '<div class="page" title="Page 1"> <div class="layoutArea"> <div class="column"> <p><span>The Norwegian sequencing centre (NSC) is a national core facility fully equipped to handle a diverse set of sequencing projects. We see an increased interest in long read technologies, in which our PacBio Single Molecule sequencing service is important. PacBio, (or SMRT) sequencing, like any other single molecule system, starts with a critical library preparation step to generate long fragment libraries of 10-20kb length. Even longer libraries can be made, but common to all library types is to start with good quality DNA of high molecular weight. Physical shearing of the DNA, targeting the desired library length, is the first critical step in the procedure. NSC has experience with different methods to fragment DNA like nebulization, enzymatic, tagmentation, covaris AFA, g-tubes (also covaris), custom syringe/needle protocols and hydroshearing. The best fragmentation method should produce a sharp fragmentation pattern close to the target length as defined by the protocol settings, wi</span><span>th as little “smear like” pattern of lower molecular length as possible. </span><span>The final step of PacBio library preparation is to size select only the longest fragments (> 7kb) as this increases the overall success rate of only sequencing the longest library fragments. It is not uncommon to loose a lot of material during this step. However, if the initial fragmentation yields sharp fragmentation patterns with minimal smear, more of the total DNA is retained in the final library. This is perhaps most important on samples were DNA amounts are limited. The optimal fragmentation method has to produce similar fragmentation pattern each time, with little deviation with sample type and input amount. We have found the new Megaruptor from Diagenode to be one of the best instruments for routine shearing of DNA to lengths of 2-40kb. It is easy to use, with walk away shearing, and the instrument does all the washes before, in between samples and after shearing without user intervention. Disposable hydropores is beneficial for lowering the contamination risk. The fact that it is so easy to use make it a good choice for laboratories with multiple users and/or when training new staff. </span></p> </div> </div> </div>', 'author' => 'Morten Skage and Dr Ave Tooming-Klunderud, Norwegian Sequencing Centre, Oslo, Norway', 'featured' => false, 'slug' => 'norwegian-sequencing-center-megaruptor', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-03-22 11:31:43', 'created' => '2015-11-26 09:21:28', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '39', 'name' => 'Megaruptor® ', 'description' => '<p><span>We will use the Megaruptor<sup>®</sup> within our PacBio<sup>®</sup> Single Molecule Sequencing workflows, especially for the construction of very big insert (>20kb) libraries.</span></p>', 'author' => 'Andrea Patrignani, Anna Bratus and Ralph Schlapbach, Functional Genomics Center Zurich (ETHZ/UZH), Switzerland', 'featured' => false, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-11-23 10:04:40', 'created' => '2015-11-23 10:03:27', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 3 => array( 'id' => '3', 'name' => 'Megaruptor - Sylke Winkler', 'description' => '<p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p>', 'author' => 'Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany', 'featured' => true, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:38:08', 'created' => '2014-10-28 23:47:30', 'ProductsTestimonial' => array( [maximum depth reached] ) ) ), 'Area' => array(), 'SafetySheet' => array() ) $country = 'US' $countries_allowed = array( (int) 0 => 'CA', (int) 1 => 'US', (int) 2 => 'IE', (int) 3 => 'GB', (int) 4 => 'DK', (int) 5 => 'NO', (int) 6 => 'SE', (int) 7 => 'FI', (int) 8 => 'NL', (int) 9 => 'BE', (int) 10 => 'LU', (int) 11 => 'FR', (int) 12 => 'DE', (int) 13 => 'CH', (int) 14 => 'AT', (int) 15 => 'ES', (int) 16 => 'IT', (int) 17 => 'PT' ) $outsource = false $other_formats = array() $edit = '' $testimonials = '<blockquote><div class="page" title="Page 1"> <div class="layoutArea"> <div class="column"> <p><span>The Norwegian sequencing centre (NSC) is a national core facility fully equipped to handle a diverse set of sequencing projects. We see an increased interest in long read technologies, in which our PacBio Single Molecule sequencing service is important. PacBio, (or SMRT) sequencing, like any other single molecule system, starts with a critical library preparation step to generate long fragment libraries of 10-20kb length. Even longer libraries can be made, but common to all library types is to start with good quality DNA of high molecular weight. Physical shearing of the DNA, targeting the desired library length, is the first critical step in the procedure. NSC has experience with different methods to fragment DNA like nebulization, enzymatic, tagmentation, covaris AFA, g-tubes (also covaris), custom syringe/needle protocols and hydroshearing. The best fragmentation method should produce a sharp fragmentation pattern close to the target length as defined by the protocol settings, wi</span><span>th as little “smear like” pattern of lower molecular length as possible. </span><span>The final step of PacBio library preparation is to size select only the longest fragments (> 7kb) as this increases the overall success rate of only sequencing the longest library fragments. It is not uncommon to loose a lot of material during this step. However, if the initial fragmentation yields sharp fragmentation patterns with minimal smear, more of the total DNA is retained in the final library. This is perhaps most important on samples were DNA amounts are limited. The optimal fragmentation method has to produce similar fragmentation pattern each time, with little deviation with sample type and input amount. We have found the new Megaruptor from Diagenode to be one of the best instruments for routine shearing of DNA to lengths of 2-40kb. It is easy to use, with walk away shearing, and the instrument does all the washes before, in between samples and after shearing without user intervention. Disposable hydropores is beneficial for lowering the contamination risk. The fact that it is so easy to use make it a good choice for laboratories with multiple users and/or when training new staff. </span></p> </div> </div> </div><cite>Morten Skage and Dr Ave Tooming-Klunderud, Norwegian Sequencing Centre, Oslo, Norway</cite></blockquote> <blockquote><p><span>We will use the Megaruptor<sup>®</sup> within our PacBio<sup>®</sup> Single Molecule Sequencing workflows, especially for the construction of very big insert (>20kb) libraries.</span></p><cite>Andrea Patrignani, Anna Bratus and Ralph Schlapbach, Functional Genomics Center Zurich (ETHZ/UZH), Switzerland</cite></blockquote> ' $featured_testimonials = '<blockquote><span class="label-green" style="margin-bottom:16px;margin-left:-22px">TESTIMONIAL</span><p>At the VIB Department of Molecular Genetics we strive to implement the best technologies in our workflows. Recent advances in long read sequencing using Oxford Nanopore technology have made the investigation of complex genomic variation in the human genome possible. A crucial step in the beginning of the library preparation is the shearing of genomic DNA, and this is where we implement the Megaruptor. The straightforward protocol is efficient, requires minimal sample handling and results in very reproducible fragment sizes. Another advantage is the consumable cost, which is lower than alternative technologies. For our applications we prefer shearing to lengths of 10 to 40kb, and for this the Megaruptor is the optimal solution.</p><cite>Wouter de Coster and the Genomic Service Facility, VIB Department of Molecular Genetics, Antwerp, Belgium</cite></blockquote> <blockquote><span class="label-green" style="margin-bottom:16px;margin-left:-22px">TESTIMONIAL</span><p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p><cite>Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany</cite></blockquote> ' $testimonial = array( 'id' => '3', 'name' => 'Megaruptor - Sylke Winkler', 'description' => '<p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p>', 'author' => 'Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany', 'featured' => true, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:38:08', 'created' => '2014-10-28 23:47:30', 'ProductsTestimonial' => array( 'id' => '24', 'product_id' => '1824', 'testimonial_id' => '3' ) ) $related_products = '<li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydro-tubes-50-pc"><img src="/img/product/shearing_technologies/C30010018%20_hydro_tubes.png" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">C30010018</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2647" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2647" id="CartAdd/2647Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2647" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydro Tubes</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydro Tubes', 'C30010018', '65', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydro Tubes', 'C30010018', '65', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydro-tubes-50-pc" data-reveal-id="cartModal-2647" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydro Tubes</h6> </div> </div> </li> <li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydropore-short-10-pc"><img src="/img/product/shearing_technologies/hydropores.jpg" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">E07010001</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2645" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2645" id="CartAdd/2645Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2645" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydropore - short</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - short', 'E07010001', '160', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - short', 'E07010001', '160', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydropore-short-10-pc" data-reveal-id="cartModal-2645" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydropore - short</h6> </div> </div> </li> <li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydropore-long-10-pc"><img src="/img/product/shearing_technologies/hydropores.jpg" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">E07010002</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2646" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2646" id="CartAdd/2646Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2646" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydropore - long</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - long', 'E07010002', '170', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - long', 'E07010002', '170', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydropore-long-10-pc" data-reveal-id="cartModal-2646" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydropore - long</h6> </div> </div> </li> ' $related = array( 'id' => '2646', 'antibody_id' => null, 'name' => 'Hydropore - long', 'description' => '<p><span style="font-weight: 400;">Hydropore-longは<strong>10 kb - 75 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010002', 'old_catalog_number' => '', 'sf_code' => 'E07010002-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '145', 'price_USD' => '170', 'price_GBP' => '130', 'price_JPY' => '22715', 'price_CNY' => '', 'price_AUD' => '425', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-long-10-pc', 'meta_title' => 'Hydropore - long', 'meta_keywords' => '', 'meta_description' => 'Hydropore - long', 'modified' => '2022-04-18 09:02:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( 'id' => '68', 'product_id' => '1824', 'related_id' => '2646' ), 'Image' => array( (int) 0 => array( 'id' => '113', 'name' => 'product/shearing_technologies/hydropores.jpg', 'alt' => 'some alt', 'modified' => '2015-06-10 17:28:55', 'created' => '2015-06-10 17:28:55', 'ProductsImage' => array( [maximum depth reached] ) ) ) ) $rrbs_service = array( (int) 0 => (int) 1894, (int) 1 => (int) 1895 ) $chipseq_service = array( (int) 0 => (int) 2683, (int) 1 => (int) 1835, (int) 2 => (int) 1836, (int) 3 => (int) 2684, (int) 4 => (int) 1838, (int) 5 => (int) 1839, (int) 6 => (int) 1856 ) $labelize = object(Closure) { } $old_catalog_number = '' $label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>' $document = array( 'id' => '783', 'name' => 'Megaruptor® DNA Shearing System User Manual v2', 'description' => '<div class="page" title="Page 4"> <div class="layoutArea"> <div class="column"> <p><span>The Megaruptor</span><span>® </span><span>was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA in the range of 2 kb - 75 kb. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p> </div> </div> </div>', 'image_id' => null, 'type' => 'Manual', 'url' => 'files/products/shearing_technology/megaruptor/Megaruptor_manual.pdf', 'slug' => 'megaruptor-manual', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-07-23 17:29:32', 'created' => '2015-07-07 11:47:45', 'ProductsDocument' => array( 'id' => '656', 'product_id' => '1824', 'document_id' => '783' ) ) $publication = array( 'id' => '2262', 'name' => 'Microevolution of Burkholderia pseudomallei during an acute infection.', 'authors' => 'Limmathurotsakul D, Holden MT, Coupland P, Price EP, Chantratita N, Wuthiekanun V, Amornchai P, Parkhill J, Peacock SJ', 'description' => 'We used whole-genome sequencing to evaluate 69 independent colonies of Burkholderia pseudomallei isolated from seven body sites of a patient with acute disseminated melioidosis. Fourteen closely related genotypes were found, providing evidence for the rapid in vivo diversification of B. pseudomallei after inoculation and systemic spread.', 'date' => '2014-09-01', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24966357', 'doi' => '', 'modified' => '2015-07-24 15:39:03', 'created' => '2015-07-24 15:39:03', 'ProductsPublication' => array( 'id' => '203', 'product_id' => '1824', 'publication_id' => '2262' ) ) $externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/24966357" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755 View::_evaluate() - CORE/Cake/View/View.php, line 971 View::_render() - CORE/Cake/View/View.php, line 933 View::render() - CORE/Cake/View/View.php, line 473 Controller::render() - CORE/Cake/Controller/Controller.php, line 963 ProductsController::slug() - APP/Controller/ProductsController.php, line 1052 ReflectionMethod::invokeArgs() - [internal], line ?? Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491 Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193 Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167 [main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: header [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL --><div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"><?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>$viewFile = '/home/website-server/www/app/View/Products/view.ctp' $dataForView = array( 'language' => 'en', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'meta_title' => 'Megaruptor', 'product' => array( 'Product' => array( 'id' => '1824', 'antibody_id' => null, 'name' => 'Megaruptor<sup>®</sup>', 'description' => '<p><span>The Megaruptor</span><sup>®</sup><span> was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA from </span><strong>2 kb - 75 kb</strong><span>. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p>', 'label1' => 'Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores', 'info1' => '<h6><img src="https://www.diagenode.com/img/product/shearing_technologies/megaruptor-fig1.png" alt="" style="display: block; margin-left: auto; margin-right: auto;" width="851" height="245" /></h6> <h6 style="text-align: center;">Validation using two different DNA sources and two different methods of analysis. <strong>A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Fragment Analyzer profiles of human genomic DNA (25 ng/μl; 200 μl/sample) sheared at different software settings of 2, 3, 5 and 8 kb. (Standard Sensitivity Large Fragment Analysis Kit ; Advanced Analytical Technologies, Inc. was used for separation and fragment sizing).</h6>', 'label2' => '', 'info2' => '', 'label3' => '', 'info3' => '', 'format' => '1 unit', 'catalog_number' => 'B06010001', 'old_catalog_number' => '', 'sf_code' => null, 'type' => 'ACC', 'search_order' => '04-undefined', 'price_EUR' => 'on request', 'price_USD' => 'on request', 'price_GBP' => 'on request', 'price_JPY' => 'on request', 'price_CNY' => 'on request', 'price_AUD' => 'on request', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => true, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'megaruptor-1-unit', 'meta_title' => 'Megaruptor', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'modified' => '2019-02-20 12:43:01', 'created' => '2015-06-29 14:08:20', 'locale' => 'eng' ), 'Antibody' => array( 'host' => '*****', 'id' => null, 'name' => null, 'description' => null, 'clonality' => null, 'isotype' => null, 'lot' => null, 'concentration' => null, 'reactivity' => null, 'type' => null, 'purity' => null, 'classification' => null, 'application_table' => null, 'storage_conditions' => null, 'storage_buffer' => null, 'precautions' => null, 'uniprot_acc' => null, 'slug' => null, 'meta_keywords' => null, 'meta_description' => null, 'modified' => null, 'created' => null, 'select_label' => null ), 'Slave' => array(), 'Group' => array(), 'Related' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ) ), 'Application' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ) ), 'Category' => array(), 'Document' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ) ), 'Feature' => array(), 'Image' => array( (int) 0 => array( [maximum depth reached] ) ), 'Promotion' => array(), 'Protocol' => array(), 'Publication' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ), (int) 3 => array( [maximum depth reached] ), (int) 4 => array( [maximum depth reached] ), (int) 5 => array( [maximum depth reached] ), (int) 6 => array( [maximum depth reached] ), (int) 7 => array( [maximum depth reached] ), (int) 8 => array( [maximum depth reached] ), (int) 9 => array( [maximum depth reached] ) ), 'Testimonial' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ), (int) 3 => array( [maximum depth reached] ) ), 'Area' => array(), 'SafetySheet' => array() ) ) $language = 'en' $meta_keywords = '' $meta_description = 'Megaruptor' $meta_title = 'Megaruptor' $product = array( 'Product' => array( 'id' => '1824', 'antibody_id' => null, 'name' => 'Megaruptor<sup>®</sup>', 'description' => '<p><span>The Megaruptor</span><sup>®</sup><span> was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA from </span><strong>2 kb - 75 kb</strong><span>. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p>', 'label1' => 'Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores', 'info1' => '<h6><img src="https://www.diagenode.com/img/product/shearing_technologies/megaruptor-fig1.png" alt="" style="display: block; margin-left: auto; margin-right: auto;" width="851" height="245" /></h6> <h6 style="text-align: center;">Validation using two different DNA sources and two different methods of analysis. <strong>A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Fragment Analyzer profiles of human genomic DNA (25 ng/μl; 200 μl/sample) sheared at different software settings of 2, 3, 5 and 8 kb. (Standard Sensitivity Large Fragment Analysis Kit ; Advanced Analytical Technologies, Inc. was used for separation and fragment sizing).</h6>', 'label2' => '', 'info2' => '', 'label3' => '', 'info3' => '', 'format' => '1 unit', 'catalog_number' => 'B06010001', 'old_catalog_number' => '', 'sf_code' => null, 'type' => 'ACC', 'search_order' => '04-undefined', 'price_EUR' => 'on request', 'price_USD' => 'on request', 'price_GBP' => 'on request', 'price_JPY' => 'on request', 'price_CNY' => 'on request', 'price_AUD' => 'on request', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => true, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'megaruptor-1-unit', 'meta_title' => 'Megaruptor', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'modified' => '2019-02-20 12:43:01', 'created' => '2015-06-29 14:08:20', 'locale' => 'eng' ), 'Antibody' => array( 'host' => '*****', 'id' => null, 'name' => null, 'description' => null, 'clonality' => null, 'isotype' => null, 'lot' => null, 'concentration' => null, 'reactivity' => null, 'type' => null, 'purity' => null, 'classification' => null, 'application_table' => null, 'storage_conditions' => null, 'storage_buffer' => null, 'precautions' => null, 'uniprot_acc' => null, 'slug' => null, 'meta_keywords' => null, 'meta_description' => null, 'modified' => null, 'created' => null, 'select_label' => null ), 'Slave' => array(), 'Group' => array(), 'Related' => array( (int) 0 => array( 'id' => '2647', 'antibody_id' => null, 'name' => 'Hydro Tubes', 'description' => '<p><span>Hydro Tubes for Megaruptor</span><sup>®</sup><span> The Microcentrifuge Tubes (0.5 ml) have been thoroughly validated for use on the Megaruptor</span><sup>®</sup><span> (B06010001) and are strongly recommended for DNA shearing applications using this instrument.</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => 'Characteristics', 'info1' => '<ul> <li>Sterile</li> <li>Natural</li> <li>Autoclavable</li> <li>Freezable</li> </ul> <p></p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '50 pc', 'catalog_number' => 'C30010018', 'old_catalog_number' => '', 'sf_code' => 'C30010018-1860', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '65', 'price_USD' => '65', 'price_GBP' => '60', 'price_JPY' => '10180', 'price_CNY' => '', 'price_AUD' => '162', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydro-tubes-50-pc', 'meta_title' => 'Hydro Tubes', 'meta_keywords' => '', 'meta_description' => 'Hydro Tubes', 'modified' => '2020-04-07 05:19:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '2645', 'antibody_id' => null, 'name' => 'Hydropore - short', 'description' => '<p><span style="font-weight: 400;">Hydropore-shortは<strong>2 kb - 9 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010001', 'old_catalog_number' => '', 'sf_code' => 'E07010001-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '135', 'price_USD' => '160', 'price_GBP' => '125', 'price_JPY' => '21150', 'price_CNY' => '', 'price_AUD' => '400', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-short-10-pc', 'meta_title' => 'Hydropore - short', 'meta_keywords' => '', 'meta_description' => 'Hydropore - short', 'modified' => '2022-04-18 09:01:09', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '2646', 'antibody_id' => null, 'name' => 'Hydropore - long', 'description' => '<p><span style="font-weight: 400;">Hydropore-longは<strong>10 kb - 75 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010002', 'old_catalog_number' => '', 'sf_code' => 'E07010002-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '145', 'price_USD' => '170', 'price_GBP' => '130', 'price_JPY' => '22715', 'price_CNY' => '', 'price_AUD' => '425', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-long-10-pc', 'meta_title' => 'Hydropore - long', 'meta_keywords' => '', 'meta_description' => 'Hydropore - long', 'modified' => '2022-04-18 09:02:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ) ), 'Application' => array( (int) 0 => array( 'id' => '13', 'position' => '10', 'parent_id' => '3', 'name' => 'DNA/RNA shearing', 'description' => '<div class="row"> <div class="small-12 medium-12 large-12 columns">In recent years, advances in Next-Generation Sequencing (NGS) have revolutionized genomics and biology. This growth has fueled demands on upstream techniques for optimal sample preparation and genomic library construction. One of the most critical aspects of optimal library preparation is the quality of the DNA to be sequenced. The DNA must first be effectively and consistently sheared into the appropriate fragment size (depending on the sequencing platform) to enable sensitive and reliable NGS results. The <strong>Bioruptor</strong><sup>®</sup> <strong>Pico</strong> and the <strong>Megaruptor</strong><sup>®</sup> provide superior sample yields, fragment size, and consistency, which are essential for Next-Generation Sequencing workflows. Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor<sup>®</sup></a>.</div> </div> <p></p> <div class="row"> <div class="small-7 medium-7 large-7 columns text-center"><img src="https://www.diagenode.com/img/applications/true-flexibility-with-br-ngs.jpg" /></div> <div class="small-5 medium-5 large-5 columns"><small><strong>Programmable DNA size distribution and high reproducibility with Bioruptor<sup>®</sup> Pico using 0.65 (panel A) or 0.1 ml (panel B) microtubes</strong>. <b>Panel A:</b> 200 bp after 13 cycles (13 sec ON/OFF) using 100 µl volume. Average size: 204; CV%:1.89%). <b>Panel B:</b> 200 bp after 20 cycles (30 sec ON/OFF) using 10 µl volume. (Average size: 215 bp; CV%: 6.6%). <b>Panel A & B:</b> peak electropherogram view. <b>Panel C & D:</b> virtual gel view.</small></div> </div> <p><br /><br /></p> <div class="row"> <div class="small-10 medium-10 large-10 columns text-center end small-offset-1"><img src="https://www.diagenode.com/img/applications/megaruptor-short-frag.jpg" /></div> <div class="small-12 medium-12 large-12 columns"><small><strong> Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores Validation using two different DNA sources and two different methods of analysis. A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Bioanalyzer profiles of human genomic DNA (20 ng/μl; 150 μl/sample) sheared at different software settings of 2 and 5 kb. Three independent experiments were run for each setting. (Agilent DNA 12000 kit was used for separation and fragment sizing).</small></div> </div> <p><br /><br /></p> <div class="row"> <div class="small-4 medium-4 large-4 columns text-center"><img src="https://www.diagenode.com/img/applications/megaruptor-long-frag.jpg" /></div> <div class="small-8 medium-8 large-8 columns"><small><strong> Demonstrated shearing to fragment sizes between 15 kb and 75 kb with Megaruptor® using long fragment size Hydropores. </strong>Image shows DNA size distribution of human genomic DNA sheared with long fragment Hydropores. DNA was analyzed by pulsed field gel electrophoresis (PFGE) in 1% agarose gel and a mean size of smears was estimated using Image Lab 4.1 software.<br /> * indicates unsheared DNA </small></div> </div>', 'in_footer' => false, 'in_menu' => true, 'online' => true, 'tabular' => false, 'slug' => 'dna-rna-shearing', 'meta_keywords' => 'DNA/RNA shearing,Bioruptor® Pico,Megaruptor®,Next-Generation Sequencing ', 'meta_description' => 'Bioruptor® Pico and the Megaruptor® provide superior sample yields, fragment size, and consistency, which are essential for Next-Generation Sequencing workflows.', 'meta_title' => 'DNA shearing & RNA shearing for Next-Generation Sequencing (NGS) | Diagenode', 'modified' => '2017-12-08 14:44:11', 'created' => '2014-10-29 12:45:41', 'ProductsApplication' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '3', 'position' => '10', 'parent_id' => null, 'name' => '次世代シーケンシング', 'description' => '<div class="row"> <div class="small-12 medium-12 large-12 columns"> <h2 style="font-size: 22px;">DNA断片化、ライブラリー調製、自動化:NGSのワンストップショップ</h2> <table class="small-12 medium-12 large-12 columns"> <tbody> <tr> <th class="small-12 medium-12 large-12 columns"> <h4>1. 断片化装置を選択してください:150 bp〜75 kbの範囲でDNAを断片化します。</h4> </th> </tr> <tr style="background-color: #ffffff;"> <td class="small-12 medium-12 large-12 columns"></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns"><a href="../p/bioruptor-pico-sonication-device"><img src="https://www.diagenode.com/img/product/shearing_technologies/bioruptor_pico.jpg" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/megaruptor2-1-unit"><img src="https://www.diagenode.com/img/product/shearing_technologies/B06010001_megaruptor2.jpg" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/bioruptor-one-sonication-device"><img src="https://www.diagenode.com/img/product/shearing_technologies/br-one-profil.png" /></a></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns">5μlまで断片化:150 bp〜2 kb<br />NGS DNAライブラリー調製およびFFPE核酸抽出に最適で、</td> <td class="small-4 medium-4 large-4 columns">2 kb〜75 kbの範囲をできます。<br />メイトペアライブラリー調製および長いフラグメントDNAシーケンシングに最適で、この軽量デスクトップデバイスで</td> <td class="small-4 medium-4 large-4 columns">20または50μlの断片化が可能です。</td> </tr> </tbody> </table> <table class="small-12 medium-12 large-12 columns"> <tbody> <tr> <th class="small-8 medium-8 large-8 columns"> <h4>2. 最適化されたライブラリー調整キットを選択してください。</h4> </th> <th class="small-4 medium-4 large-4 columns"> <h4>3. ライブラリー前処理自動化を選択して、比類のないデータ再現性を実感</h4> </th> </tr> <tr style="background-color: #ffffff;"> <td class="small-12 medium-12 large-12 columns"></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns"><a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns"><img src="https://www.diagenode.com/img/product/kits/microPlex_library_preparation.png" style="display: block; margin-left: auto; margin-right: auto;" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/ideal-library-preparation-kit-x24-incl-index-primer-set-1-24-rxns"><img src="https://www.diagenode.com/img/product/kits/box_kit.jpg" style="display: block; margin-left: auto; margin-right: auto;" height="173" width="250" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/sx-8g-ip-star-compact-automated-system-1-unit"><img src="https://www.diagenode.com/img/product/automation/B03000002%20_ipstar_compact.png" style="display: block; margin-left: auto; margin-right: auto;" /></a></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">50pgの低入力:MicroPlex Library Preparation Kit</td> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">5ng以上:iDeal Library Preparation Kit</td> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">Achieve great NGS data easily</td> </tr> </tbody> </table> </div> </div> <blockquote> <div class="row"> <div class="small-12 medium-12 large-12 columns"><span class="label" style="margin-bottom: 16px; margin-left: -22px; font-size: 15px;">DiagenodeがNGS研究にぴったりなプロバイダーである理由</span> <p>Diagenodeは15年以上もエピジェネティクス研究に専念、専門としています。 ChIP研究クロマチン用のユニークな断片化システムの開発から始まり、 専門知識を活かし、5μlのせん断体積まで可能で、NGS DNAライブラリーの調製に最適な最先端DNA断片化装置の開発にたどり着きました。 我々は以来、ChIP-seq、Methyl-seq、NGSライブラリー調製用キットを研究開発し、業界をリードする免疫沈降研究と同様に、ライブラリー調製を自動化および完結させる独自の自動化システムを開発にも成功しました。</p> <ul> <li>信頼されるせん断装置</li> <li>様々なインプットからのライブラリ作成キット</li> <li>独自の自動化デバイス</li> </ul> </div> </div> </blockquote> <div class="row"> <div class="small-12 columns"> <ul class="accordion" data-accordion=""> <li class="accordion-navigation"><a href="#panel1a">次世代シーケンシングへの理解とその専門知識</a> <div id="panel1a" class="content"> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p><strong>次世代シーケンシング (NGS)</strong> )は、著しいスケールとハイスループットでシーケンシングを行い、1日に数十億もの塩基生成を可能にします。 NGSのハイスループットは迅速でありながら正確で、再現性のあるデータセットを実現し、さらにシーケンシング費用を削減します。 NGSは、ゲノムシーケンシング、ゲノム再シーケンシング、デノボシーケンシング、トランスクリプトームシーケンシング、その他にDNA-タンパク質相互作用の検出やエピゲノムなどを示します。 指数関数的に増加するシーケンシングデータの需要は、計算分析の障害や解釈、データストレージなどの課題を解決します。</p> <p>アプリケーションおよび出発物質に応じて、数百万から数十億の鋳型DNA分子を大規模に並行してシーケンシングすることが可能です。その為に、異なる化学物質を使用するいくつかの市販のNGSプラットフォームを利用することができます。 NGSプラットフォームの種類によっては、事前準備とライブラリー作成が必要です。</p> <p>NGSにとっても、特にデータ処理と分析に関した大きな課題はあります。第3世代技術はゲノミクス研究にさらに革命を起こすであろうと大きく期待されています。</p> </div> </div> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <p><strong>NGS アプリケーション</strong></p> <ul> <li>全ゲノム配列決定</li> <li>デノボシーケンシング</li> <li>標的配列</li> <li>Exomeシーケンシング</li> <li>トランスクリプトーム配列決定</li> <li>ゲノム配列決定</li> <li>ミトコンドリア配列決定</li> <li>DNA-タンパク質相互作用(ChIP-seq</li> <li>バリアント検出</li> <li>ゲノム仕上げ</li> </ul> </div> <div class="small-6 medium-6 large-6 columns"> <p><strong>研究分野におけるNGS:</strong></p> <ul> <li>腫瘍学</li> <li>リプロダクティブ・ヘルス</li> <li>法医学ゲノミクス</li> <li>アグリゲノミックス</li> <li>複雑な病気</li> <li>微生物ゲノミクス</li> <li>食品・環境ゲノミクス</li> <li>創薬ゲノミクス - パーソナライズド・メディカル</li> </ul> </div> <div class="small-12 medium-12 large-12 columns"> <p><strong>NGSの用語</strong></p> <dl> <dt>リード(読み取り)</dt> <dd>この装置から得られた連続した単一のストレッチ</dd> <dt>断片リード</dt> <dd>フラグメントライブラリからの読み込み。 シーケンシングプラットフォームに応じて、読み取りは通常約100〜300bp。</dd> <dt>断片ペアエンドリード</dt> <dd>断片ライブラリーからDNA断片の各末端2つの読み取り。</dd> <dt>メイトペアリード</dt> <dd>大きなDNA断片(通常は予め定義されたサイズ範囲)の各末端から2つの読み取り。</dd> <dt>カバレッジ(例)</dt> <dd>30×適用範囲とは、参照ゲノム中の各塩基対が平均30回の読み取りを示す。</dd> </dl> </div> </div> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <h2>NGSプラットフォーム</h2> <h3><a href="http://www.illumina.com" target="_blank">イルミナ</a></h3> <p>イルミナは、クローン的に増幅された鋳型DNA(クラスター)上に位置する、蛍光標識された可逆的鎖ターミネーターヌクレオチドを用いた配列別合成技術を使用。 DNAクラスターは、ガラスフローセルの表面上に固定化され、 ワークフローは、4つのヌクレオチド(それぞれ異なる蛍光色素で標識された)の組み込み、4色イメージング、色素や末端基の切断、取り込み、イメージングなどを繰り返します。フローセルは大規模な並列配列決定を受ける。 この方法により、単一蛍光標識されたヌクレオチドの制御添加によるモノヌクレオチドのエラーを回避する可能性があります。 読み取りの長さは、通常約100〜150 bpです。</p> <h3><a href="http://www.lifetechnologies.com" target="_blank">イオン トレント</a></h3> <p>イオントレントは、半導体技術チップを用いて、合成中にヌクレオチドを取り込む際に放出されたプロトンを検出します。 これは、イオン球粒子と呼ばれるビーズの表面にエマルションPCR(emPCR)を使用し、リンクされた特定のアダプターを用いてDNA断片を増幅します。 各ビーズは1種類のDNA断片で覆われていて、異なるDNA断片を有するビーズは次いで、チップの陽子感知ウェル内に配置されます。 チップには一度に4つのヌクレオチドのうちの1つが浸水し、このプロセスは異なるヌクレオチドで15秒ごとに繰り返されます。 配列決定の間に4つの塩基の各々が1つずつ導入されます、組み込みの場合はプロトンが放出され、電圧信号が取り込みに比例して検出されます。.</p> <h3><a href="http://www.pacificbiosciences.com" target="_blank">パシフィック バイオサイエンス</a></h3> <p>パシフィックバイオサイエンスでは、20kbを超える塩基対の読み取りも、単一分子リアルタイム(SMRT)シーケンシングによる構造および細胞タイプの変化を観察することができます。 このプラットフォームでは、超長鎖二本鎖DNA(dsDNA)断片が、Megaruptor(登録商標)のようなDiagenode装置を用いたランダムシアリングまたは目的の標的領域の増幅によって生成されます。 SMRTbellライブラリーは、ユニバーサルヘアピンアダプターをDNA断片の各末端に連結することによって生成します。 サイズ選択条件による洗浄ステップの後、配列決定プライマーをSMRTbellテンプレートにアニーリングし、鋳型DNAに結合したDNAポリメラーゼを含む配列決定を、蛍光標識ヌクレオチドの存在下で開始。 各塩基が取り込まれると、異なる蛍光のパルスをリアルタイムで検出します。</p> <h3><a href="https://nanoporetech.com" target="_blank">オックスフォード ナノポア</a></h3> <p>Oxford Nanoporeは、単一のDNA分子配列決定に基づく技術を開発します。その技術により生物学的分子、すなわちDNAが一群の電気抵抗性高分子膜として位置するナノスケールの孔(ナノ細孔)またはその近くを通過し、イオン電流が変化します。 この変化に関する情報は、例えば4つのヌクレオチド(AまたはG r CまたはT)ならびに修飾されたヌクレオチドすべてを区別することによって分子情報に訳されます。 シーケンシングミニオンデバイスのフローセルは、数百個のナノポアチャネルのセンサアレイを含みます。 DNAサンプルは、Diagenode社のMegaruptor(登録商標)を用いてランダムシアリングによって生成され得る超長鎖DNAフラグメントが必要です。</p> <h3><a href="http://www.lifetechnologies.com/be/en/home/life-science/sequencing/next-generation-sequencing/solid-next-generation-sequencing.html" target="_blank">SOLiD</a></h3> <p>SOLiDは、ユニークな化学作用により、何千という個々のDNA分子の同時配列決定を可能にします。 それは、アダプター対ライブラリーのフラグメントが適切で、せん断されたゲノムDNAへのアダプターのライゲーションによるライブラリー作製から始まります。 次のステップでは、エマルジョンPCR(emPCR)を実施して、ビーズの表面上の個々の鋳型DNA分子をクローン的に増幅。 emPCRでは、個々の鋳型DNAをPCR試薬と混合し、水中油型エマルジョン内の疎水性シェルで囲まれた水性液滴内のプライマーコートビーズを、配列決定のためにロードするスライドガラスの表面にランダムに付着。 この技術は、シークエンシングプライマーへのライゲーションで競合する4つの蛍光標識されたジ塩基プローブのセットを使用します。</p> <h3><a href="http://454.com/products/technology.asp" target="_blank">454</a></h3> <p>454は、大規模並列パイロシーケンシングを利用しています。 始めに全ゲノムDNAまたは標的遺伝子断片の300〜800bp断片のライブラリー調製します。 次に、DNAフラグメントへのアダプターの付着および単一のDNA鎖の分離。 その後アダプターに連結されたDNAフラグメントをエマルジョンベースのクローン増幅(emPCR)で処理し、DNAライブラリーフラグメントをミクロンサイズのビーズ上に配置します。 各DNA結合ビーズを光ファイバーチップ上のウェルに入れ、器具に挿入します。 4つのDNAヌクレオチドは、配列決定操作中に固定された順序で連続して加えられ、並行して配列決定されます。</p> </div> </div> </div> </li> </ul> </div> </div>', 'in_footer' => true, 'in_menu' => true, 'online' => true, 'tabular' => true, 'slug' => 'next-generation-sequencing', 'meta_keywords' => 'Next-generation sequencing,NGS,Whole genome sequencing,NGS platforms,DNA/RNA shearing', 'meta_description' => 'Diagenode offers kits and DNA/RNA shearing technology prior to next-generation sequencing, many Next-generation sequencing platforms require preparation of the DNA.', 'meta_title' => 'Next-generation sequencing (NGS) Applications and Methods | Diagenode', 'modified' => '2018-07-26 05:24:29', 'created' => '2015-04-01 22:47:04', 'ProductsApplication' => array( [maximum depth reached] ) ) ), 'Category' => array(), 'Document' => array( (int) 0 => array( 'id' => '14', 'name' => 'Improvements in Hydrodynamic Shearing Facilitates 20-50 kb True Mate Pair Library Construction', 'description' => '<p><span>The Megaruptor<sup>®</sup> has been used by several sequencing centers to produce high-quality small insert fragment libraries for Illumina and PacBio platforms. David Mead and colleagues from Lucigen<sup>®</sup> have used the Megaruptor<sup>®</sup> to prepare 20-50 kb inserts for a new mate-pair library technology that is >70% efficient. </span></p>', 'image_id' => null, 'type' => 'Poster', 'url' => 'files/posters/Mead_et_al_Lucigen_Diagenode_Poster.pdf', 'slug' => 'mead-et-al-lucigen-diagenode-poster', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-09-29 13:42:27', 'created' => '2015-07-03 16:05:15', 'ProductsDocument' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '783', 'name' => 'Megaruptor® DNA Shearing System User Manual v2', 'description' => '<div class="page" title="Page 4"> <div class="layoutArea"> <div class="column"> <p><span>The Megaruptor</span><span>® </span><span>was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA in the range of 2 kb - 75 kb. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p> </div> </div> </div>', 'image_id' => null, 'type' => 'Manual', 'url' => 'files/products/shearing_technology/megaruptor/Megaruptor_manual.pdf', 'slug' => 'megaruptor-manual', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-07-23 17:29:32', 'created' => '2015-07-07 11:47:45', 'ProductsDocument' => array( [maximum depth reached] ) ) ), 'Feature' => array(), 'Image' => array( (int) 0 => array( 'id' => '139', 'name' => 'product/shearing_technologies/B06010001_megaruptor.jpg', 'alt' => 'megaruptor', 'modified' => '2015-07-23 17:39:35', 'created' => '2015-06-10 17:28:55', 'ProductsImage' => array( [maximum depth reached] ) ) ), 'Promotion' => array(), 'Protocol' => array(), 'Publication' => array( (int) 0 => array( 'id' => '4018', 'name' => 'Complete Genome Sequence of the Type Strain Pectobacterium punjabenseSS95, Isolated from a Potato Plant with Blackleg Symptoms.', 'authors' => 'Sarfraz, S and Oulghazi, S and Cigna, J and Sahi, ST and Riaz, K andTufail, MR and Fayyaz, A and Naveed, K and Hameed, A and Lopez-Roques, Cand Vandecasteele, C and Faure, D', 'description' => '<p>Pectobacterium punjabense is a newly described species causing blackleg disease in potato plants. Therefore, by the combination of long (Oxford Nanopore Technologies, MinION) and short (Illumina MiSeq) reads, we sequenced the complete genome of P. punjabense SS95T, which contains a circular chromosome of 4.793 Mb with a GC content of 50.7\%.</p>', 'date' => '2020-08-06', 'pmid' => 'http://www.pubmed.gov/32763925', 'doi' => '10.1128/MRA.00420-20.', 'modified' => '2020-12-16 17:37:51', 'created' => '2020-10-12 14:54:59', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '4001', 'name' => 'Sizing, stabilising, and cloning repeat-expansions for gene targeting constructs.', 'authors' => 'Nair RR, Tibbit C, Thompson D, McLeod R, Nakhuda A, Simon MM, Baloh RH, Fisher EMC, Isaacs AM, Cunningham TJ', 'description' => '<p>Aberrant microsatellite repeat-expansions at specific loci within the human genome cause several distinct, heritable, and predominantly neurological, disorders. Creating models for these diseases poses a challenge, due to the instability of such repeats in bacterial vectors, especially with large repeat expansions. Designing constructs for more precise genome engineering projects, such as engineering knock-in mice, proves a greater challenge still, since these unstable repeats require numerous cloning steps in order to introduce homology arms or selection cassettes. Here, we report our efforts to clone a large hexanucleotide repeat in the C9orf72 gene, originating from within a BAC construct, derived from a C9orf72-ALS patient. We provide detailed methods for efficient repeat sizing and growth conditions in bacteria to facilitate repeat retention during growth and sub-culturing. We report that sub-cloning into a linear vector dramatically improves stability, but is dependent on the relative orientation of DNA replication through the repeat, consistent with previous studies. We envisage the findings presented here provide a relatively straightforward route to maintaining large-range microsatellite repeat-expansions, for efficient cloning into vectors.</p>', 'date' => '2020-07-25', 'pmid' => 'http://www.pubmed.gov/32721467', 'doi' => '10.1016/j.ymeth.2020.07.007', 'modified' => '2020-09-01 14:39:41', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '3980', 'name' => 'Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants.', 'authors' => 'Xu Z, Pu X, Gao R, Demurtas OC, Fleck SJ, Richter M, He C, Ji A, Sun W, Kong J, Hu K, Ren F, Song J, Wang Z, Gao T, Xiong C, Yu H, Xin T, Albert VA, Giuliano G, Chen S, Song J', 'description' => '<p>BACKGROUND: Plants have evolved a panoply of specialized metabolites that increase their environmental fitness. Two examples are caffeine, a purine psychotropic alkaloid, and crocins, a group of glycosylated apocarotenoid pigments. Both classes of compounds are found in a handful of distantly related plant genera (Coffea, Camellia, Paullinia, and Ilex for caffeine; Crocus, Buddleja, and Gardenia for crocins) wherein they presumably evolved through convergent evolution. The closely related Coffea and Gardenia genera belong to the Rubiaceae family and synthesize, respectively, caffeine and crocins in their fruits. RESULTS: Here, we report a chromosomal-level genome assembly of Gardenia jasminoides, a crocin-producing species, obtained using Oxford Nanopore sequencing and Hi-C technology. Through genomic and functional assays, we completely deciphered for the first time in any plant the dedicated pathway of crocin biosynthesis. Through comparative analyses with Coffea canephora and other eudicot genomes, we show that Coffea caffeine synthases and the first dedicated gene in the Gardenia crocin pathway, GjCCD4a, evolved through recent tandem gene duplications in the two different genera, respectively. In contrast, genes encoding later steps of the Gardenia crocin pathway, ALDH and UGT, evolved through more ancient gene duplications and were presumably recruited into the crocin biosynthetic pathway only after the evolution of the GjCCD4a gene. CONCLUSIONS: This study shows duplication-based divergent evolution within the coffee family (Rubiaceae) of two characteristic secondary metabolic pathways, caffeine and crocin biosynthesis, from a common ancestor that possessed neither complete pathway. These findings provide significant insights on the role of tandem duplications in the evolution of plant specialized metabolism.</p>', 'date' => '2020-06-18', 'pmid' => 'http://www.pubmed.gov/32552824', 'doi' => '10.1186/s12915-020-00795-3', 'modified' => '2020-09-01 15:27:14', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 3 => array( 'id' => '4002', 'name' => 'Nanopore Sequencing and Its Clinical Applications.', 'authors' => 'Sun X, Song L, Yang W, Zhang L, Liu M, Li X, Tian G, Wang W', 'description' => '<p>Nanopore sequencing is a method for determining the order and modifications of DNA/RNA nucleotides by detecting the electric current variations when DNA/RNA oligonucleotides pass through the nanometer-sized hole (nanopore). Nanopore-based DNA analysis techniques have been commercialized by Oxford Nanopore Technologies, NabSys, and Sequenom, and widely used in scientific researches recently including human genomics, cancer, metagenomics, plant sciences, etc., moreover, it also has potential applications in the field of healthcare due to its fast turn-around time, portable and real-time data analysis. Those features make it a promising technology for the point-of-care testing (POCT) and its potential clinical applications are briefly discussed in this chapter.</p>', 'date' => '2020-01-01', 'pmid' => 'http://www.pubmed.gov/32710311', 'doi' => '10.1007/978-1-0716-0904-0_2,', 'modified' => '2020-09-01 14:39:13', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 4 => array( 'id' => '3045', 'name' => 'Condition-dependent co-regulation of genomic clusters of virulence factors in the grapevine trunk pathogen Neofusicoccum parvum', 'authors' => 'Massonnet M. et al.', 'description' => '<p>The ascomycete Neofusicoccum parvum, one of the causal agents of Botryosphaeria dieback, is a destructive wood-infecting fungus and a serious threat to grape production worldwide. The capability of colonizing woody tissue combined with the secretion of phytotoxic compounds is thought to underlie its pathogenicity and virulence. Here, we describe the repertoire of virulence factors and their transcriptional dynamics as the fungus feeds on different substrates and colonizes the woody stem. We assembled and annotated a highly contiguous genome using single molecule real-time DNA sequencing. Transcriptome profiling by RNA-sequencing determined the genome-wide patterns of expression of virulence factors both in vitro (potato dextrose agar or medium amended with grape wood as substrate) and in planta. Pairwise statistical testing of differential expression followed by co-expression network analysis revealed that physically clustered genes coding for putative virulence functions were induced depending on substrate or stage of plant infection. Co-expressed gene clusters were significantly enriched not only in genes associated with secondary metabolism, but also with cell wall degradation, suggesting that dynamic co-regulation of transcriptional networks contribute to multiple aspects of N. parvum virulence. In most of the co-expressed clusters, all genes shared at least a common motif in their promoter region indicative of co-regulation by the same transcription factor. Co-expression analysis also identified chromatin regulators with correlated expression with inducible clusters of virulence factors, suggesting a complex, multi-layered regulation of the virulence repertoire of N. parvum.</p>', 'date' => '2016-09-08', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27608421', 'doi' => '', 'modified' => '2016-10-10 10:53:20', 'created' => '2016-10-10 10:53:20', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 5 => array( 'id' => '2839', 'name' => 'Complete genome of Staphylococcus aureus Tager 104 provides evidence of its relation to modern systemic hospital-acquired strains', 'authors' => 'Richard W. DavisIV, Andrew D. Brannen, Mohammad J. Hossain, Scott Monsma, Paul E. Bock, Matthias Nahrendorf, David Mead, Michael Lodes, Mark R. Liles and Peter Panizzi', 'description' => '<section xmlns="" xmlns:meta="http://www.springer.com/app/meta" class="Abstract" id="Abs1" lang="en"> <div class="js-CollapseSection"> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec1"> <h5 class="Heading">Background</h5> <p id="Par1" class="Para"><em xmlns="" class="EmphasisTypeItalic">Staphylococcus aureus</em> (<em xmlns="" class="EmphasisTypeItalic">S. aureus</em>) infections range in severity due to expression of certain virulence factors encoded on mobile genetic elements (MGE). As such, characterization of these MGE, as well as single nucleotide polymorphisms, is of high clinical and microbiological importance. To understand the evolution of these dangerous pathogens, it is paramount to define reference strains that may predate MGE acquisition. One such candidate is <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> Tager 104, a previously uncharacterized strain isolated from a patient with impetigo in 1947.</p> </div> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec2"> <h5 class="Heading">Results</h5> <p id="Par2" class="Para">We show here that <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> Tager 104 can survive in the bloodstream and infect naïve organs. We also demonstrate a procedure to construct and validate the assembly of <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> genomes, using Tager 104 as a proof-of-concept. In so doing, we bridged confounding gap regions that limited our initial attempts to close this 2.82 Mb genome, through integration of data from Illumina Nextera paired-end, PacBio RS, and Lucigen NxSeq mate-pair libraries. Furthermore, we provide independent confirmation of our segmental arrangement of the Tager 104 genome by the sole use of Lucigen NxSeq libraries filled by paired-end MiSeq reads and alignment with SPAdes software. Genomic analysis of Tager 104 revealed limited MGE, and a νSaβ island configuration that is reminiscent of other hospital acquired <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> genomes.</p> </div> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec3"> <h5 class="Heading">Conclusions</h5> <p id="Par3" class="Para">Tager 104 represents an early-branching ancestor of certain hospital-acquired strains. Combined with its earlier isolation date and limited content of MGE, Tager 104 can serve as a viable reference for future comparative genome studies.</p> </div> </div> </section>', 'date' => '2016-03-03', 'pmid' => 'http://pubmed.gov/26940863', 'doi' => '10.1186/s12864-016-2433-8', 'modified' => '2016-03-09 10:13:09', 'created' => '2016-03-09 10:13:09', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 6 => array( 'id' => '2953', 'name' => 'Complete Genome Sequences of Four Escherichia coli ST95 Isolates from Bloodstream Infections', 'authors' => 'Craig M. Stephens, Jeffrey M. Skerker, Manraj S. Sekhon, Adam P. Arkin, Lee W. Rileyd', 'description' => '<p>Finished genome sequences are presented for four Escherichia coli strains isolated from bloodstream infections at San Francisco General Hospital. These strains provide reference sequences for four major fimH-identified sublineages within the multilocus sequence type (MLST) ST95 group, and provide insights into pathogenicity and differential antimicrobial susceptibility within this group.</p>', 'date' => '2015-11-05', 'pmid' => 'http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4645194/', 'doi' => '10.1128/genomeA.01241-15', 'modified' => '2016-06-13 15:50:25', 'created' => '2016-06-13 15:50:25', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 7 => array( 'id' => '2372', 'name' => 'Diversification of bacterial genome content through distinct mechanisms over different timescales.', 'authors' => 'Croucher NJ, Coupland PG, Stevenson AE, Callendrello A, Bentley SD, Hanage WP', 'description' => 'Bacterial populations often consist of multiple co-circulating lineages. Determining how such population structures arise requires understanding what drives bacterial diversification. Using 616 systematically sampled genomes, we show that Streptococcus pneumoniae lineages are typically characterized by combinations of infrequently transferred stable genomic islands: those moving primarily through transformation, along with integrative and conjugative elements and phage-related chromosomal islands. The only lineage containing extensive unique sequence corresponds to a set of atypical unencapsulated isolates that may represent a distinct species. However, prophage content is highly variable even within lineages, suggesting frequent horizontal transmission that would necessitate rapidly diversifying anti-phage mechanisms to prevent these viruses sweeping through populations. Correspondingly, two loci encoding Type I restriction-modification systems able to change their specificity over short timescales through intragenomic recombination are ubiquitous across the collection. Hence short-term pneumococcal variation is characterized by movement of phage and intragenomic rearrangements, with the slower transfer of stable loci distinguishing lineages.', 'date' => '2014-11-19', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25407023', 'doi' => '', 'modified' => '2015-07-24 15:39:04', 'created' => '2015-07-24 15:39:04', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 8 => array( 'id' => '2838', 'name' => 'Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance', 'authors' => 'Mark R Davies, Matthew T Holden, Paul Coupland, Jonathan H K Chen, Carola Venturini, Timothy C Barnett, Nouri L Ben Zakour, Herman Tse, Gordon Dougan, Kwok-Yung Yuen & Mark J Walker', 'description' => '<p><span>A scarlet fever outbreak began in mainland China and Hong Kong in 2011 (refs. </span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref1" title="Chen, M. et al. Outbreak of scarlet fever associated with emm12 type group A Streptococcus in 2011 in Shanghai, China. Pediatr. Infect. Dis. J. 31, e158-e162 (2012)." id="ref-link-1">1</a><span>–</span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref6" title="Yang, P. et al. Characteristics of group A Streptococcus strains circulating during scarlet fever epidemic, Beijing, China, 2011. Emerg. Infect. Dis. 19, 909-915 (2013)." id="ref-link-2">6</a><span>). Macrolide- and tetracycline-resistant </span><i>Streptococcus pyogenes emm</i><span>12 isolates represent the majority of clinical cases. Recently, we identified two mobile genetic elements that were closely associated with </span><i>emm</i><span>12 outbreak isolates: the integrative and conjugative element ICE-</span><i>emm</i><span>12, encoding genes for tetracycline and macrolide resistance, and prophage </span><span class="mb">Φ</span><span>HKU.vir, encoding the superantigens SSA and SpeC, as well as the DNase Spd1 (ref. </span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref4" title="Tse, H. et al. Molecular characterization of the 2011 Hong Kong scarlet fever outbreak. J. Infect. Dis. 206, 341-351 (2012)." id="ref-link-3">4</a><span>). Here we sequenced the genomes of 141 </span><i>emm</i><span>12 isolates, including 132 isolated in Hong Kong between 2005 and 2011. We found that the introduction of several ICE-</span><i>emm</i><span>12 variants, </span><span class="mb">Φ</span><span>HKU.vir and a new prophage, </span><span class="mb">Φ</span><span>HKU.ssa, occurred in three distinct </span><i>emm</i><span>12 lineages late in the twentieth century. Acquisition of </span><i>ssa</i><span> and transposable elements encoding multidrug resistance genes triggered the expansion of scarlet fever–associated </span><i>emm</i><span>12 lineages in Hong Kong. The occurrence of multidrug-resistant </span><i>ssa</i><span>-harboring scarlet fever strains should prompt heightened surveillance within China and abroad for the dissemination of these mobile genetic elements.</span></p>', 'date' => '2014-11-17', 'pmid' => 'http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html', 'doi' => '10.1038/ng.3147', 'modified' => '2016-03-08 14:33:49', 'created' => '2016-03-08 14:33:21', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 9 => array( 'id' => '2262', 'name' => 'Microevolution of Burkholderia pseudomallei during an acute infection.', 'authors' => 'Limmathurotsakul D, Holden MT, Coupland P, Price EP, Chantratita N, Wuthiekanun V, Amornchai P, Parkhill J, Peacock SJ', 'description' => 'We used whole-genome sequencing to evaluate 69 independent colonies of Burkholderia pseudomallei isolated from seven body sites of a patient with acute disseminated melioidosis. Fourteen closely related genotypes were found, providing evidence for the rapid in vivo diversification of B. pseudomallei after inoculation and systemic spread.', 'date' => '2014-09-01', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24966357', 'doi' => '', 'modified' => '2015-07-24 15:39:03', 'created' => '2015-07-24 15:39:03', 'ProductsPublication' => array( [maximum depth reached] ) ) ), 'Testimonial' => array( (int) 0 => array( 'id' => '57', 'name' => 'VIB - Megaruptor', 'description' => '<p>At the VIB Department of Molecular Genetics we strive to implement the best technologies in our workflows. Recent advances in long read sequencing using Oxford Nanopore technology have made the investigation of complex genomic variation in the human genome possible. A crucial step in the beginning of the library preparation is the shearing of genomic DNA, and this is where we implement the Megaruptor. The straightforward protocol is efficient, requires minimal sample handling and results in very reproducible fragment sizes. Another advantage is the consumable cost, which is lower than alternative technologies. For our applications we prefer shearing to lengths of 10 to 40kb, and for this the Megaruptor is the optimal solution.</p>', 'author' => 'Wouter de Coster and the Genomic Service Facility, VIB Department of Molecular Genetics, Antwerp, Belgium', 'featured' => true, 'slug' => 'vib-antwerp', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:35:57', 'created' => '2016-07-06 10:35:57', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '42', 'name' => 'Norwegian Sequencing Center - Megaruptor', 'description' => '<div class="page" title="Page 1"> <div class="layoutArea"> <div class="column"> <p><span>The Norwegian sequencing centre (NSC) is a national core facility fully equipped to handle a diverse set of sequencing projects. We see an increased interest in long read technologies, in which our PacBio Single Molecule sequencing service is important. PacBio, (or SMRT) sequencing, like any other single molecule system, starts with a critical library preparation step to generate long fragment libraries of 10-20kb length. Even longer libraries can be made, but common to all library types is to start with good quality DNA of high molecular weight. Physical shearing of the DNA, targeting the desired library length, is the first critical step in the procedure. NSC has experience with different methods to fragment DNA like nebulization, enzymatic, tagmentation, covaris AFA, g-tubes (also covaris), custom syringe/needle protocols and hydroshearing. The best fragmentation method should produce a sharp fragmentation pattern close to the target length as defined by the protocol settings, wi</span><span>th as little “smear like” pattern of lower molecular length as possible. </span><span>The final step of PacBio library preparation is to size select only the longest fragments (> 7kb) as this increases the overall success rate of only sequencing the longest library fragments. It is not uncommon to loose a lot of material during this step. However, if the initial fragmentation yields sharp fragmentation patterns with minimal smear, more of the total DNA is retained in the final library. This is perhaps most important on samples were DNA amounts are limited. The optimal fragmentation method has to produce similar fragmentation pattern each time, with little deviation with sample type and input amount. We have found the new Megaruptor from Diagenode to be one of the best instruments for routine shearing of DNA to lengths of 2-40kb. It is easy to use, with walk away shearing, and the instrument does all the washes before, in between samples and after shearing without user intervention. Disposable hydropores is beneficial for lowering the contamination risk. The fact that it is so easy to use make it a good choice for laboratories with multiple users and/or when training new staff. </span></p> </div> </div> </div>', 'author' => 'Morten Skage and Dr Ave Tooming-Klunderud, Norwegian Sequencing Centre, Oslo, Norway', 'featured' => false, 'slug' => 'norwegian-sequencing-center-megaruptor', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-03-22 11:31:43', 'created' => '2015-11-26 09:21:28', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '39', 'name' => 'Megaruptor® ', 'description' => '<p><span>We will use the Megaruptor<sup>®</sup> within our PacBio<sup>®</sup> Single Molecule Sequencing workflows, especially for the construction of very big insert (>20kb) libraries.</span></p>', 'author' => 'Andrea Patrignani, Anna Bratus and Ralph Schlapbach, Functional Genomics Center Zurich (ETHZ/UZH), Switzerland', 'featured' => false, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-11-23 10:04:40', 'created' => '2015-11-23 10:03:27', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 3 => array( 'id' => '3', 'name' => 'Megaruptor - Sylke Winkler', 'description' => '<p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p>', 'author' => 'Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany', 'featured' => true, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:38:08', 'created' => '2014-10-28 23:47:30', 'ProductsTestimonial' => array( [maximum depth reached] ) ) ), 'Area' => array(), 'SafetySheet' => array() ) $country = 'US' $countries_allowed = array( (int) 0 => 'CA', (int) 1 => 'US', (int) 2 => 'IE', (int) 3 => 'GB', (int) 4 => 'DK', (int) 5 => 'NO', (int) 6 => 'SE', (int) 7 => 'FI', (int) 8 => 'NL', (int) 9 => 'BE', (int) 10 => 'LU', (int) 11 => 'FR', (int) 12 => 'DE', (int) 13 => 'CH', (int) 14 => 'AT', (int) 15 => 'ES', (int) 16 => 'IT', (int) 17 => 'PT' ) $outsource = false $other_formats = array() $edit = '' $testimonials = '<blockquote><div class="page" title="Page 1"> <div class="layoutArea"> <div class="column"> <p><span>The Norwegian sequencing centre (NSC) is a national core facility fully equipped to handle a diverse set of sequencing projects. We see an increased interest in long read technologies, in which our PacBio Single Molecule sequencing service is important. PacBio, (or SMRT) sequencing, like any other single molecule system, starts with a critical library preparation step to generate long fragment libraries of 10-20kb length. Even longer libraries can be made, but common to all library types is to start with good quality DNA of high molecular weight. Physical shearing of the DNA, targeting the desired library length, is the first critical step in the procedure. NSC has experience with different methods to fragment DNA like nebulization, enzymatic, tagmentation, covaris AFA, g-tubes (also covaris), custom syringe/needle protocols and hydroshearing. The best fragmentation method should produce a sharp fragmentation pattern close to the target length as defined by the protocol settings, wi</span><span>th as little “smear like” pattern of lower molecular length as possible. </span><span>The final step of PacBio library preparation is to size select only the longest fragments (> 7kb) as this increases the overall success rate of only sequencing the longest library fragments. It is not uncommon to loose a lot of material during this step. However, if the initial fragmentation yields sharp fragmentation patterns with minimal smear, more of the total DNA is retained in the final library. This is perhaps most important on samples were DNA amounts are limited. The optimal fragmentation method has to produce similar fragmentation pattern each time, with little deviation with sample type and input amount. We have found the new Megaruptor from Diagenode to be one of the best instruments for routine shearing of DNA to lengths of 2-40kb. It is easy to use, with walk away shearing, and the instrument does all the washes before, in between samples and after shearing without user intervention. Disposable hydropores is beneficial for lowering the contamination risk. The fact that it is so easy to use make it a good choice for laboratories with multiple users and/or when training new staff. </span></p> </div> </div> </div><cite>Morten Skage and Dr Ave Tooming-Klunderud, Norwegian Sequencing Centre, Oslo, Norway</cite></blockquote> <blockquote><p><span>We will use the Megaruptor<sup>®</sup> within our PacBio<sup>®</sup> Single Molecule Sequencing workflows, especially for the construction of very big insert (>20kb) libraries.</span></p><cite>Andrea Patrignani, Anna Bratus and Ralph Schlapbach, Functional Genomics Center Zurich (ETHZ/UZH), Switzerland</cite></blockquote> ' $featured_testimonials = '<blockquote><span class="label-green" style="margin-bottom:16px;margin-left:-22px">TESTIMONIAL</span><p>At the VIB Department of Molecular Genetics we strive to implement the best technologies in our workflows. Recent advances in long read sequencing using Oxford Nanopore technology have made the investigation of complex genomic variation in the human genome possible. A crucial step in the beginning of the library preparation is the shearing of genomic DNA, and this is where we implement the Megaruptor. The straightforward protocol is efficient, requires minimal sample handling and results in very reproducible fragment sizes. Another advantage is the consumable cost, which is lower than alternative technologies. For our applications we prefer shearing to lengths of 10 to 40kb, and for this the Megaruptor is the optimal solution.</p><cite>Wouter de Coster and the Genomic Service Facility, VIB Department of Molecular Genetics, Antwerp, Belgium</cite></blockquote> <blockquote><span class="label-green" style="margin-bottom:16px;margin-left:-22px">TESTIMONIAL</span><p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p><cite>Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany</cite></blockquote> ' $testimonial = array( 'id' => '3', 'name' => 'Megaruptor - Sylke Winkler', 'description' => '<p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p>', 'author' => 'Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany', 'featured' => true, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:38:08', 'created' => '2014-10-28 23:47:30', 'ProductsTestimonial' => array( 'id' => '24', 'product_id' => '1824', 'testimonial_id' => '3' ) ) $related_products = '<li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydro-tubes-50-pc"><img src="/img/product/shearing_technologies/C30010018%20_hydro_tubes.png" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">C30010018</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2647" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2647" id="CartAdd/2647Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2647" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydro Tubes</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydro Tubes', 'C30010018', '65', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydro Tubes', 'C30010018', '65', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydro-tubes-50-pc" data-reveal-id="cartModal-2647" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydro Tubes</h6> </div> </div> </li> <li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydropore-short-10-pc"><img src="/img/product/shearing_technologies/hydropores.jpg" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">E07010001</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2645" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2645" id="CartAdd/2645Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2645" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydropore - short</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - short', 'E07010001', '160', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - short', 'E07010001', '160', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydropore-short-10-pc" data-reveal-id="cartModal-2645" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydropore - short</h6> </div> </div> </li> <li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydropore-long-10-pc"><img src="/img/product/shearing_technologies/hydropores.jpg" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">E07010002</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2646" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2646" id="CartAdd/2646Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2646" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydropore - long</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - long', 'E07010002', '170', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - long', 'E07010002', '170', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydropore-long-10-pc" data-reveal-id="cartModal-2646" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydropore - long</h6> </div> </div> </li> ' $related = array( 'id' => '2646', 'antibody_id' => null, 'name' => 'Hydropore - long', 'description' => '<p><span style="font-weight: 400;">Hydropore-longは<strong>10 kb - 75 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010002', 'old_catalog_number' => '', 'sf_code' => 'E07010002-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '145', 'price_USD' => '170', 'price_GBP' => '130', 'price_JPY' => '22715', 'price_CNY' => '', 'price_AUD' => '425', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-long-10-pc', 'meta_title' => 'Hydropore - long', 'meta_keywords' => '', 'meta_description' => 'Hydropore - long', 'modified' => '2022-04-18 09:02:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( 'id' => '68', 'product_id' => '1824', 'related_id' => '2646' ), 'Image' => array( (int) 0 => array( 'id' => '113', 'name' => 'product/shearing_technologies/hydropores.jpg', 'alt' => 'some alt', 'modified' => '2015-06-10 17:28:55', 'created' => '2015-06-10 17:28:55', 'ProductsImage' => array( [maximum depth reached] ) ) ) ) $rrbs_service = array( (int) 0 => (int) 1894, (int) 1 => (int) 1895 ) $chipseq_service = array( (int) 0 => (int) 2683, (int) 1 => (int) 1835, (int) 2 => (int) 1836, (int) 3 => (int) 2684, (int) 4 => (int) 1838, (int) 5 => (int) 1839, (int) 6 => (int) 1856 ) $labelize = object(Closure) { } $old_catalog_number = '' $label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>' $document = array( 'id' => '783', 'name' => 'Megaruptor® DNA Shearing System User Manual v2', 'description' => '<div class="page" title="Page 4"> <div class="layoutArea"> <div class="column"> <p><span>The Megaruptor</span><span>® </span><span>was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA in the range of 2 kb - 75 kb. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p> </div> </div> </div>', 'image_id' => null, 'type' => 'Manual', 'url' => 'files/products/shearing_technology/megaruptor/Megaruptor_manual.pdf', 'slug' => 'megaruptor-manual', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-07-23 17:29:32', 'created' => '2015-07-07 11:47:45', 'ProductsDocument' => array( 'id' => '656', 'product_id' => '1824', 'document_id' => '783' ) ) $publication = array( 'id' => '2262', 'name' => 'Microevolution of Burkholderia pseudomallei during an acute infection.', 'authors' => 'Limmathurotsakul D, Holden MT, Coupland P, Price EP, Chantratita N, Wuthiekanun V, Amornchai P, Parkhill J, Peacock SJ', 'description' => 'We used whole-genome sequencing to evaluate 69 independent colonies of Burkholderia pseudomallei isolated from seven body sites of a patient with acute disseminated melioidosis. Fourteen closely related genotypes were found, providing evidence for the rapid in vivo diversification of B. pseudomallei after inoculation and systemic spread.', 'date' => '2014-09-01', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24966357', 'doi' => '', 'modified' => '2015-07-24 15:39:03', 'created' => '2015-07-24 15:39:03', 'ProductsPublication' => array( 'id' => '203', 'product_id' => '1824', 'publication_id' => '2262' ) ) $externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/24966357" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755 View::_evaluate() - CORE/Cake/View/View.php, line 971 View::_render() - CORE/Cake/View/View.php, line 933 View::render() - CORE/Cake/View/View.php, line 473 Controller::render() - CORE/Cake/Controller/Controller.php, line 963 ProductsController::slug() - APP/Controller/ProductsController.php, line 1052 ReflectionMethod::invokeArgs() - [internal], line ?? Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491 Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193 Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167 [main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: message [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL --><div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"><?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>$viewFile = '/home/website-server/www/app/View/Products/view.ctp' $dataForView = array( 'language' => 'en', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'meta_title' => 'Megaruptor', 'product' => array( 'Product' => array( 'id' => '1824', 'antibody_id' => null, 'name' => 'Megaruptor<sup>®</sup>', 'description' => '<p><span>The Megaruptor</span><sup>®</sup><span> was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA from </span><strong>2 kb - 75 kb</strong><span>. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p>', 'label1' => 'Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores', 'info1' => '<h6><img src="https://www.diagenode.com/img/product/shearing_technologies/megaruptor-fig1.png" alt="" style="display: block; margin-left: auto; margin-right: auto;" width="851" height="245" /></h6> <h6 style="text-align: center;">Validation using two different DNA sources and two different methods of analysis. <strong>A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Fragment Analyzer profiles of human genomic DNA (25 ng/μl; 200 μl/sample) sheared at different software settings of 2, 3, 5 and 8 kb. (Standard Sensitivity Large Fragment Analysis Kit ; Advanced Analytical Technologies, Inc. was used for separation and fragment sizing).</h6>', 'label2' => '', 'info2' => '', 'label3' => '', 'info3' => '', 'format' => '1 unit', 'catalog_number' => 'B06010001', 'old_catalog_number' => '', 'sf_code' => null, 'type' => 'ACC', 'search_order' => '04-undefined', 'price_EUR' => 'on request', 'price_USD' => 'on request', 'price_GBP' => 'on request', 'price_JPY' => 'on request', 'price_CNY' => 'on request', 'price_AUD' => 'on request', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => true, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'megaruptor-1-unit', 'meta_title' => 'Megaruptor', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'modified' => '2019-02-20 12:43:01', 'created' => '2015-06-29 14:08:20', 'locale' => 'eng' ), 'Antibody' => array( 'host' => '*****', 'id' => null, 'name' => null, 'description' => null, 'clonality' => null, 'isotype' => null, 'lot' => null, 'concentration' => null, 'reactivity' => null, 'type' => null, 'purity' => null, 'classification' => null, 'application_table' => null, 'storage_conditions' => null, 'storage_buffer' => null, 'precautions' => null, 'uniprot_acc' => null, 'slug' => null, 'meta_keywords' => null, 'meta_description' => null, 'modified' => null, 'created' => null, 'select_label' => null ), 'Slave' => array(), 'Group' => array(), 'Related' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ) ), 'Application' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ) ), 'Category' => array(), 'Document' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ) ), 'Feature' => array(), 'Image' => array( (int) 0 => array( [maximum depth reached] ) ), 'Promotion' => array(), 'Protocol' => array(), 'Publication' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ), (int) 3 => array( [maximum depth reached] ), (int) 4 => array( [maximum depth reached] ), (int) 5 => array( [maximum depth reached] ), (int) 6 => array( [maximum depth reached] ), (int) 7 => array( [maximum depth reached] ), (int) 8 => array( [maximum depth reached] ), (int) 9 => array( [maximum depth reached] ) ), 'Testimonial' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ), (int) 3 => array( [maximum depth reached] ) ), 'Area' => array(), 'SafetySheet' => array() ) ) $language = 'en' $meta_keywords = '' $meta_description = 'Megaruptor' $meta_title = 'Megaruptor' $product = array( 'Product' => array( 'id' => '1824', 'antibody_id' => null, 'name' => 'Megaruptor<sup>®</sup>', 'description' => '<p><span>The Megaruptor</span><sup>®</sup><span> was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA from </span><strong>2 kb - 75 kb</strong><span>. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p>', 'label1' => 'Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores', 'info1' => '<h6><img src="https://www.diagenode.com/img/product/shearing_technologies/megaruptor-fig1.png" alt="" style="display: block; margin-left: auto; margin-right: auto;" width="851" height="245" /></h6> <h6 style="text-align: center;">Validation using two different DNA sources and two different methods of analysis. <strong>A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Fragment Analyzer profiles of human genomic DNA (25 ng/μl; 200 μl/sample) sheared at different software settings of 2, 3, 5 and 8 kb. (Standard Sensitivity Large Fragment Analysis Kit ; Advanced Analytical Technologies, Inc. was used for separation and fragment sizing).</h6>', 'label2' => '', 'info2' => '', 'label3' => '', 'info3' => '', 'format' => '1 unit', 'catalog_number' => 'B06010001', 'old_catalog_number' => '', 'sf_code' => null, 'type' => 'ACC', 'search_order' => '04-undefined', 'price_EUR' => 'on request', 'price_USD' => 'on request', 'price_GBP' => 'on request', 'price_JPY' => 'on request', 'price_CNY' => 'on request', 'price_AUD' => 'on request', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => true, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'megaruptor-1-unit', 'meta_title' => 'Megaruptor', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'modified' => '2019-02-20 12:43:01', 'created' => '2015-06-29 14:08:20', 'locale' => 'eng' ), 'Antibody' => array( 'host' => '*****', 'id' => null, 'name' => null, 'description' => null, 'clonality' => null, 'isotype' => null, 'lot' => null, 'concentration' => null, 'reactivity' => null, 'type' => null, 'purity' => null, 'classification' => null, 'application_table' => null, 'storage_conditions' => null, 'storage_buffer' => null, 'precautions' => null, 'uniprot_acc' => null, 'slug' => null, 'meta_keywords' => null, 'meta_description' => null, 'modified' => null, 'created' => null, 'select_label' => null ), 'Slave' => array(), 'Group' => array(), 'Related' => array( (int) 0 => array( 'id' => '2647', 'antibody_id' => null, 'name' => 'Hydro Tubes', 'description' => '<p><span>Hydro Tubes for Megaruptor</span><sup>®</sup><span> The Microcentrifuge Tubes (0.5 ml) have been thoroughly validated for use on the Megaruptor</span><sup>®</sup><span> (B06010001) and are strongly recommended for DNA shearing applications using this instrument.</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => 'Characteristics', 'info1' => '<ul> <li>Sterile</li> <li>Natural</li> <li>Autoclavable</li> <li>Freezable</li> </ul> <p></p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '50 pc', 'catalog_number' => 'C30010018', 'old_catalog_number' => '', 'sf_code' => 'C30010018-1860', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '65', 'price_USD' => '65', 'price_GBP' => '60', 'price_JPY' => '10180', 'price_CNY' => '', 'price_AUD' => '162', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydro-tubes-50-pc', 'meta_title' => 'Hydro Tubes', 'meta_keywords' => '', 'meta_description' => 'Hydro Tubes', 'modified' => '2020-04-07 05:19:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '2645', 'antibody_id' => null, 'name' => 'Hydropore - short', 'description' => '<p><span style="font-weight: 400;">Hydropore-shortは<strong>2 kb - 9 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010001', 'old_catalog_number' => '', 'sf_code' => 'E07010001-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '135', 'price_USD' => '160', 'price_GBP' => '125', 'price_JPY' => '21150', 'price_CNY' => '', 'price_AUD' => '400', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-short-10-pc', 'meta_title' => 'Hydropore - short', 'meta_keywords' => '', 'meta_description' => 'Hydropore - short', 'modified' => '2022-04-18 09:01:09', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '2646', 'antibody_id' => null, 'name' => 'Hydropore - long', 'description' => '<p><span style="font-weight: 400;">Hydropore-longは<strong>10 kb - 75 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010002', 'old_catalog_number' => '', 'sf_code' => 'E07010002-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '145', 'price_USD' => '170', 'price_GBP' => '130', 'price_JPY' => '22715', 'price_CNY' => '', 'price_AUD' => '425', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-long-10-pc', 'meta_title' => 'Hydropore - long', 'meta_keywords' => '', 'meta_description' => 'Hydropore - long', 'modified' => '2022-04-18 09:02:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ) ), 'Application' => array( (int) 0 => array( 'id' => '13', 'position' => '10', 'parent_id' => '3', 'name' => 'DNA/RNA shearing', 'description' => '<div class="row"> <div class="small-12 medium-12 large-12 columns">In recent years, advances in Next-Generation Sequencing (NGS) have revolutionized genomics and biology. This growth has fueled demands on upstream techniques for optimal sample preparation and genomic library construction. One of the most critical aspects of optimal library preparation is the quality of the DNA to be sequenced. The DNA must first be effectively and consistently sheared into the appropriate fragment size (depending on the sequencing platform) to enable sensitive and reliable NGS results. The <strong>Bioruptor</strong><sup>®</sup> <strong>Pico</strong> and the <strong>Megaruptor</strong><sup>®</sup> provide superior sample yields, fragment size, and consistency, which are essential for Next-Generation Sequencing workflows. Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor<sup>®</sup></a>.</div> </div> <p></p> <div class="row"> <div class="small-7 medium-7 large-7 columns text-center"><img src="https://www.diagenode.com/img/applications/true-flexibility-with-br-ngs.jpg" /></div> <div class="small-5 medium-5 large-5 columns"><small><strong>Programmable DNA size distribution and high reproducibility with Bioruptor<sup>®</sup> Pico using 0.65 (panel A) or 0.1 ml (panel B) microtubes</strong>. <b>Panel A:</b> 200 bp after 13 cycles (13 sec ON/OFF) using 100 µl volume. Average size: 204; CV%:1.89%). <b>Panel B:</b> 200 bp after 20 cycles (30 sec ON/OFF) using 10 µl volume. (Average size: 215 bp; CV%: 6.6%). <b>Panel A & B:</b> peak electropherogram view. <b>Panel C & D:</b> virtual gel view.</small></div> </div> <p><br /><br /></p> <div class="row"> <div class="small-10 medium-10 large-10 columns text-center end small-offset-1"><img src="https://www.diagenode.com/img/applications/megaruptor-short-frag.jpg" /></div> <div class="small-12 medium-12 large-12 columns"><small><strong> Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores Validation using two different DNA sources and two different methods of analysis. A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Bioanalyzer profiles of human genomic DNA (20 ng/μl; 150 μl/sample) sheared at different software settings of 2 and 5 kb. Three independent experiments were run for each setting. (Agilent DNA 12000 kit was used for separation and fragment sizing).</small></div> </div> <p><br /><br /></p> <div class="row"> <div class="small-4 medium-4 large-4 columns text-center"><img src="https://www.diagenode.com/img/applications/megaruptor-long-frag.jpg" /></div> <div class="small-8 medium-8 large-8 columns"><small><strong> Demonstrated shearing to fragment sizes between 15 kb and 75 kb with Megaruptor® using long fragment size Hydropores. </strong>Image shows DNA size distribution of human genomic DNA sheared with long fragment Hydropores. DNA was analyzed by pulsed field gel electrophoresis (PFGE) in 1% agarose gel and a mean size of smears was estimated using Image Lab 4.1 software.<br /> * indicates unsheared DNA </small></div> </div>', 'in_footer' => false, 'in_menu' => true, 'online' => true, 'tabular' => false, 'slug' => 'dna-rna-shearing', 'meta_keywords' => 'DNA/RNA shearing,Bioruptor® Pico,Megaruptor®,Next-Generation Sequencing ', 'meta_description' => 'Bioruptor® Pico and the Megaruptor® provide superior sample yields, fragment size, and consistency, which are essential for Next-Generation Sequencing workflows.', 'meta_title' => 'DNA shearing & RNA shearing for Next-Generation Sequencing (NGS) | Diagenode', 'modified' => '2017-12-08 14:44:11', 'created' => '2014-10-29 12:45:41', 'ProductsApplication' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '3', 'position' => '10', 'parent_id' => null, 'name' => '次世代シーケンシング', 'description' => '<div class="row"> <div class="small-12 medium-12 large-12 columns"> <h2 style="font-size: 22px;">DNA断片化、ライブラリー調製、自動化:NGSのワンストップショップ</h2> <table class="small-12 medium-12 large-12 columns"> <tbody> <tr> <th class="small-12 medium-12 large-12 columns"> <h4>1. 断片化装置を選択してください:150 bp〜75 kbの範囲でDNAを断片化します。</h4> </th> </tr> <tr style="background-color: #ffffff;"> <td class="small-12 medium-12 large-12 columns"></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns"><a href="../p/bioruptor-pico-sonication-device"><img src="https://www.diagenode.com/img/product/shearing_technologies/bioruptor_pico.jpg" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/megaruptor2-1-unit"><img src="https://www.diagenode.com/img/product/shearing_technologies/B06010001_megaruptor2.jpg" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/bioruptor-one-sonication-device"><img src="https://www.diagenode.com/img/product/shearing_technologies/br-one-profil.png" /></a></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns">5μlまで断片化:150 bp〜2 kb<br />NGS DNAライブラリー調製およびFFPE核酸抽出に最適で、</td> <td class="small-4 medium-4 large-4 columns">2 kb〜75 kbの範囲をできます。<br />メイトペアライブラリー調製および長いフラグメントDNAシーケンシングに最適で、この軽量デスクトップデバイスで</td> <td class="small-4 medium-4 large-4 columns">20または50μlの断片化が可能です。</td> </tr> </tbody> </table> <table class="small-12 medium-12 large-12 columns"> <tbody> <tr> <th class="small-8 medium-8 large-8 columns"> <h4>2. 最適化されたライブラリー調整キットを選択してください。</h4> </th> <th class="small-4 medium-4 large-4 columns"> <h4>3. ライブラリー前処理自動化を選択して、比類のないデータ再現性を実感</h4> </th> </tr> <tr style="background-color: #ffffff;"> <td class="small-12 medium-12 large-12 columns"></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns"><a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns"><img src="https://www.diagenode.com/img/product/kits/microPlex_library_preparation.png" style="display: block; margin-left: auto; margin-right: auto;" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/ideal-library-preparation-kit-x24-incl-index-primer-set-1-24-rxns"><img src="https://www.diagenode.com/img/product/kits/box_kit.jpg" style="display: block; margin-left: auto; margin-right: auto;" height="173" width="250" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/sx-8g-ip-star-compact-automated-system-1-unit"><img src="https://www.diagenode.com/img/product/automation/B03000002%20_ipstar_compact.png" style="display: block; margin-left: auto; margin-right: auto;" /></a></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">50pgの低入力:MicroPlex Library Preparation Kit</td> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">5ng以上:iDeal Library Preparation Kit</td> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">Achieve great NGS data easily</td> </tr> </tbody> </table> </div> </div> <blockquote> <div class="row"> <div class="small-12 medium-12 large-12 columns"><span class="label" style="margin-bottom: 16px; margin-left: -22px; font-size: 15px;">DiagenodeがNGS研究にぴったりなプロバイダーである理由</span> <p>Diagenodeは15年以上もエピジェネティクス研究に専念、専門としています。 ChIP研究クロマチン用のユニークな断片化システムの開発から始まり、 専門知識を活かし、5μlのせん断体積まで可能で、NGS DNAライブラリーの調製に最適な最先端DNA断片化装置の開発にたどり着きました。 我々は以来、ChIP-seq、Methyl-seq、NGSライブラリー調製用キットを研究開発し、業界をリードする免疫沈降研究と同様に、ライブラリー調製を自動化および完結させる独自の自動化システムを開発にも成功しました。</p> <ul> <li>信頼されるせん断装置</li> <li>様々なインプットからのライブラリ作成キット</li> <li>独自の自動化デバイス</li> </ul> </div> </div> </blockquote> <div class="row"> <div class="small-12 columns"> <ul class="accordion" data-accordion=""> <li class="accordion-navigation"><a href="#panel1a">次世代シーケンシングへの理解とその専門知識</a> <div id="panel1a" class="content"> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p><strong>次世代シーケンシング (NGS)</strong> )は、著しいスケールとハイスループットでシーケンシングを行い、1日に数十億もの塩基生成を可能にします。 NGSのハイスループットは迅速でありながら正確で、再現性のあるデータセットを実現し、さらにシーケンシング費用を削減します。 NGSは、ゲノムシーケンシング、ゲノム再シーケンシング、デノボシーケンシング、トランスクリプトームシーケンシング、その他にDNA-タンパク質相互作用の検出やエピゲノムなどを示します。 指数関数的に増加するシーケンシングデータの需要は、計算分析の障害や解釈、データストレージなどの課題を解決します。</p> <p>アプリケーションおよび出発物質に応じて、数百万から数十億の鋳型DNA分子を大規模に並行してシーケンシングすることが可能です。その為に、異なる化学物質を使用するいくつかの市販のNGSプラットフォームを利用することができます。 NGSプラットフォームの種類によっては、事前準備とライブラリー作成が必要です。</p> <p>NGSにとっても、特にデータ処理と分析に関した大きな課題はあります。第3世代技術はゲノミクス研究にさらに革命を起こすであろうと大きく期待されています。</p> </div> </div> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <p><strong>NGS アプリケーション</strong></p> <ul> <li>全ゲノム配列決定</li> <li>デノボシーケンシング</li> <li>標的配列</li> <li>Exomeシーケンシング</li> <li>トランスクリプトーム配列決定</li> <li>ゲノム配列決定</li> <li>ミトコンドリア配列決定</li> <li>DNA-タンパク質相互作用(ChIP-seq</li> <li>バリアント検出</li> <li>ゲノム仕上げ</li> </ul> </div> <div class="small-6 medium-6 large-6 columns"> <p><strong>研究分野におけるNGS:</strong></p> <ul> <li>腫瘍学</li> <li>リプロダクティブ・ヘルス</li> <li>法医学ゲノミクス</li> <li>アグリゲノミックス</li> <li>複雑な病気</li> <li>微生物ゲノミクス</li> <li>食品・環境ゲノミクス</li> <li>創薬ゲノミクス - パーソナライズド・メディカル</li> </ul> </div> <div class="small-12 medium-12 large-12 columns"> <p><strong>NGSの用語</strong></p> <dl> <dt>リード(読み取り)</dt> <dd>この装置から得られた連続した単一のストレッチ</dd> <dt>断片リード</dt> <dd>フラグメントライブラリからの読み込み。 シーケンシングプラットフォームに応じて、読み取りは通常約100〜300bp。</dd> <dt>断片ペアエンドリード</dt> <dd>断片ライブラリーからDNA断片の各末端2つの読み取り。</dd> <dt>メイトペアリード</dt> <dd>大きなDNA断片(通常は予め定義されたサイズ範囲)の各末端から2つの読み取り。</dd> <dt>カバレッジ(例)</dt> <dd>30×適用範囲とは、参照ゲノム中の各塩基対が平均30回の読み取りを示す。</dd> </dl> </div> </div> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <h2>NGSプラットフォーム</h2> <h3><a href="http://www.illumina.com" target="_blank">イルミナ</a></h3> <p>イルミナは、クローン的に増幅された鋳型DNA(クラスター)上に位置する、蛍光標識された可逆的鎖ターミネーターヌクレオチドを用いた配列別合成技術を使用。 DNAクラスターは、ガラスフローセルの表面上に固定化され、 ワークフローは、4つのヌクレオチド(それぞれ異なる蛍光色素で標識された)の組み込み、4色イメージング、色素や末端基の切断、取り込み、イメージングなどを繰り返します。フローセルは大規模な並列配列決定を受ける。 この方法により、単一蛍光標識されたヌクレオチドの制御添加によるモノヌクレオチドのエラーを回避する可能性があります。 読み取りの長さは、通常約100〜150 bpです。</p> <h3><a href="http://www.lifetechnologies.com" target="_blank">イオン トレント</a></h3> <p>イオントレントは、半導体技術チップを用いて、合成中にヌクレオチドを取り込む際に放出されたプロトンを検出します。 これは、イオン球粒子と呼ばれるビーズの表面にエマルションPCR(emPCR)を使用し、リンクされた特定のアダプターを用いてDNA断片を増幅します。 各ビーズは1種類のDNA断片で覆われていて、異なるDNA断片を有するビーズは次いで、チップの陽子感知ウェル内に配置されます。 チップには一度に4つのヌクレオチドのうちの1つが浸水し、このプロセスは異なるヌクレオチドで15秒ごとに繰り返されます。 配列決定の間に4つの塩基の各々が1つずつ導入されます、組み込みの場合はプロトンが放出され、電圧信号が取り込みに比例して検出されます。.</p> <h3><a href="http://www.pacificbiosciences.com" target="_blank">パシフィック バイオサイエンス</a></h3> <p>パシフィックバイオサイエンスでは、20kbを超える塩基対の読み取りも、単一分子リアルタイム(SMRT)シーケンシングによる構造および細胞タイプの変化を観察することができます。 このプラットフォームでは、超長鎖二本鎖DNA(dsDNA)断片が、Megaruptor(登録商標)のようなDiagenode装置を用いたランダムシアリングまたは目的の標的領域の増幅によって生成されます。 SMRTbellライブラリーは、ユニバーサルヘアピンアダプターをDNA断片の各末端に連結することによって生成します。 サイズ選択条件による洗浄ステップの後、配列決定プライマーをSMRTbellテンプレートにアニーリングし、鋳型DNAに結合したDNAポリメラーゼを含む配列決定を、蛍光標識ヌクレオチドの存在下で開始。 各塩基が取り込まれると、異なる蛍光のパルスをリアルタイムで検出します。</p> <h3><a href="https://nanoporetech.com" target="_blank">オックスフォード ナノポア</a></h3> <p>Oxford Nanoporeは、単一のDNA分子配列決定に基づく技術を開発します。その技術により生物学的分子、すなわちDNAが一群の電気抵抗性高分子膜として位置するナノスケールの孔(ナノ細孔)またはその近くを通過し、イオン電流が変化します。 この変化に関する情報は、例えば4つのヌクレオチド(AまたはG r CまたはT)ならびに修飾されたヌクレオチドすべてを区別することによって分子情報に訳されます。 シーケンシングミニオンデバイスのフローセルは、数百個のナノポアチャネルのセンサアレイを含みます。 DNAサンプルは、Diagenode社のMegaruptor(登録商標)を用いてランダムシアリングによって生成され得る超長鎖DNAフラグメントが必要です。</p> <h3><a href="http://www.lifetechnologies.com/be/en/home/life-science/sequencing/next-generation-sequencing/solid-next-generation-sequencing.html" target="_blank">SOLiD</a></h3> <p>SOLiDは、ユニークな化学作用により、何千という個々のDNA分子の同時配列決定を可能にします。 それは、アダプター対ライブラリーのフラグメントが適切で、せん断されたゲノムDNAへのアダプターのライゲーションによるライブラリー作製から始まります。 次のステップでは、エマルジョンPCR(emPCR)を実施して、ビーズの表面上の個々の鋳型DNA分子をクローン的に増幅。 emPCRでは、個々の鋳型DNAをPCR試薬と混合し、水中油型エマルジョン内の疎水性シェルで囲まれた水性液滴内のプライマーコートビーズを、配列決定のためにロードするスライドガラスの表面にランダムに付着。 この技術は、シークエンシングプライマーへのライゲーションで競合する4つの蛍光標識されたジ塩基プローブのセットを使用します。</p> <h3><a href="http://454.com/products/technology.asp" target="_blank">454</a></h3> <p>454は、大規模並列パイロシーケンシングを利用しています。 始めに全ゲノムDNAまたは標的遺伝子断片の300〜800bp断片のライブラリー調製します。 次に、DNAフラグメントへのアダプターの付着および単一のDNA鎖の分離。 その後アダプターに連結されたDNAフラグメントをエマルジョンベースのクローン増幅(emPCR)で処理し、DNAライブラリーフラグメントをミクロンサイズのビーズ上に配置します。 各DNA結合ビーズを光ファイバーチップ上のウェルに入れ、器具に挿入します。 4つのDNAヌクレオチドは、配列決定操作中に固定された順序で連続して加えられ、並行して配列決定されます。</p> </div> </div> </div> </li> </ul> </div> </div>', 'in_footer' => true, 'in_menu' => true, 'online' => true, 'tabular' => true, 'slug' => 'next-generation-sequencing', 'meta_keywords' => 'Next-generation sequencing,NGS,Whole genome sequencing,NGS platforms,DNA/RNA shearing', 'meta_description' => 'Diagenode offers kits and DNA/RNA shearing technology prior to next-generation sequencing, many Next-generation sequencing platforms require preparation of the DNA.', 'meta_title' => 'Next-generation sequencing (NGS) Applications and Methods | Diagenode', 'modified' => '2018-07-26 05:24:29', 'created' => '2015-04-01 22:47:04', 'ProductsApplication' => array( [maximum depth reached] ) ) ), 'Category' => array(), 'Document' => array( (int) 0 => array( 'id' => '14', 'name' => 'Improvements in Hydrodynamic Shearing Facilitates 20-50 kb True Mate Pair Library Construction', 'description' => '<p><span>The Megaruptor<sup>®</sup> has been used by several sequencing centers to produce high-quality small insert fragment libraries for Illumina and PacBio platforms. David Mead and colleagues from Lucigen<sup>®</sup> have used the Megaruptor<sup>®</sup> to prepare 20-50 kb inserts for a new mate-pair library technology that is >70% efficient. </span></p>', 'image_id' => null, 'type' => 'Poster', 'url' => 'files/posters/Mead_et_al_Lucigen_Diagenode_Poster.pdf', 'slug' => 'mead-et-al-lucigen-diagenode-poster', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-09-29 13:42:27', 'created' => '2015-07-03 16:05:15', 'ProductsDocument' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '783', 'name' => 'Megaruptor® DNA Shearing System User Manual v2', 'description' => '<div class="page" title="Page 4"> <div class="layoutArea"> <div class="column"> <p><span>The Megaruptor</span><span>® </span><span>was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA in the range of 2 kb - 75 kb. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p> </div> </div> </div>', 'image_id' => null, 'type' => 'Manual', 'url' => 'files/products/shearing_technology/megaruptor/Megaruptor_manual.pdf', 'slug' => 'megaruptor-manual', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-07-23 17:29:32', 'created' => '2015-07-07 11:47:45', 'ProductsDocument' => array( [maximum depth reached] ) ) ), 'Feature' => array(), 'Image' => array( (int) 0 => array( 'id' => '139', 'name' => 'product/shearing_technologies/B06010001_megaruptor.jpg', 'alt' => 'megaruptor', 'modified' => '2015-07-23 17:39:35', 'created' => '2015-06-10 17:28:55', 'ProductsImage' => array( [maximum depth reached] ) ) ), 'Promotion' => array(), 'Protocol' => array(), 'Publication' => array( (int) 0 => array( 'id' => '4018', 'name' => 'Complete Genome Sequence of the Type Strain Pectobacterium punjabenseSS95, Isolated from a Potato Plant with Blackleg Symptoms.', 'authors' => 'Sarfraz, S and Oulghazi, S and Cigna, J and Sahi, ST and Riaz, K andTufail, MR and Fayyaz, A and Naveed, K and Hameed, A and Lopez-Roques, Cand Vandecasteele, C and Faure, D', 'description' => '<p>Pectobacterium punjabense is a newly described species causing blackleg disease in potato plants. Therefore, by the combination of long (Oxford Nanopore Technologies, MinION) and short (Illumina MiSeq) reads, we sequenced the complete genome of P. punjabense SS95T, which contains a circular chromosome of 4.793 Mb with a GC content of 50.7\%.</p>', 'date' => '2020-08-06', 'pmid' => 'http://www.pubmed.gov/32763925', 'doi' => '10.1128/MRA.00420-20.', 'modified' => '2020-12-16 17:37:51', 'created' => '2020-10-12 14:54:59', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '4001', 'name' => 'Sizing, stabilising, and cloning repeat-expansions for gene targeting constructs.', 'authors' => 'Nair RR, Tibbit C, Thompson D, McLeod R, Nakhuda A, Simon MM, Baloh RH, Fisher EMC, Isaacs AM, Cunningham TJ', 'description' => '<p>Aberrant microsatellite repeat-expansions at specific loci within the human genome cause several distinct, heritable, and predominantly neurological, disorders. Creating models for these diseases poses a challenge, due to the instability of such repeats in bacterial vectors, especially with large repeat expansions. Designing constructs for more precise genome engineering projects, such as engineering knock-in mice, proves a greater challenge still, since these unstable repeats require numerous cloning steps in order to introduce homology arms or selection cassettes. Here, we report our efforts to clone a large hexanucleotide repeat in the C9orf72 gene, originating from within a BAC construct, derived from a C9orf72-ALS patient. We provide detailed methods for efficient repeat sizing and growth conditions in bacteria to facilitate repeat retention during growth and sub-culturing. We report that sub-cloning into a linear vector dramatically improves stability, but is dependent on the relative orientation of DNA replication through the repeat, consistent with previous studies. We envisage the findings presented here provide a relatively straightforward route to maintaining large-range microsatellite repeat-expansions, for efficient cloning into vectors.</p>', 'date' => '2020-07-25', 'pmid' => 'http://www.pubmed.gov/32721467', 'doi' => '10.1016/j.ymeth.2020.07.007', 'modified' => '2020-09-01 14:39:41', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '3980', 'name' => 'Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants.', 'authors' => 'Xu Z, Pu X, Gao R, Demurtas OC, Fleck SJ, Richter M, He C, Ji A, Sun W, Kong J, Hu K, Ren F, Song J, Wang Z, Gao T, Xiong C, Yu H, Xin T, Albert VA, Giuliano G, Chen S, Song J', 'description' => '<p>BACKGROUND: Plants have evolved a panoply of specialized metabolites that increase their environmental fitness. Two examples are caffeine, a purine psychotropic alkaloid, and crocins, a group of glycosylated apocarotenoid pigments. Both classes of compounds are found in a handful of distantly related plant genera (Coffea, Camellia, Paullinia, and Ilex for caffeine; Crocus, Buddleja, and Gardenia for crocins) wherein they presumably evolved through convergent evolution. The closely related Coffea and Gardenia genera belong to the Rubiaceae family and synthesize, respectively, caffeine and crocins in their fruits. RESULTS: Here, we report a chromosomal-level genome assembly of Gardenia jasminoides, a crocin-producing species, obtained using Oxford Nanopore sequencing and Hi-C technology. Through genomic and functional assays, we completely deciphered for the first time in any plant the dedicated pathway of crocin biosynthesis. Through comparative analyses with Coffea canephora and other eudicot genomes, we show that Coffea caffeine synthases and the first dedicated gene in the Gardenia crocin pathway, GjCCD4a, evolved through recent tandem gene duplications in the two different genera, respectively. In contrast, genes encoding later steps of the Gardenia crocin pathway, ALDH and UGT, evolved through more ancient gene duplications and were presumably recruited into the crocin biosynthetic pathway only after the evolution of the GjCCD4a gene. CONCLUSIONS: This study shows duplication-based divergent evolution within the coffee family (Rubiaceae) of two characteristic secondary metabolic pathways, caffeine and crocin biosynthesis, from a common ancestor that possessed neither complete pathway. These findings provide significant insights on the role of tandem duplications in the evolution of plant specialized metabolism.</p>', 'date' => '2020-06-18', 'pmid' => 'http://www.pubmed.gov/32552824', 'doi' => '10.1186/s12915-020-00795-3', 'modified' => '2020-09-01 15:27:14', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 3 => array( 'id' => '4002', 'name' => 'Nanopore Sequencing and Its Clinical Applications.', 'authors' => 'Sun X, Song L, Yang W, Zhang L, Liu M, Li X, Tian G, Wang W', 'description' => '<p>Nanopore sequencing is a method for determining the order and modifications of DNA/RNA nucleotides by detecting the electric current variations when DNA/RNA oligonucleotides pass through the nanometer-sized hole (nanopore). Nanopore-based DNA analysis techniques have been commercialized by Oxford Nanopore Technologies, NabSys, and Sequenom, and widely used in scientific researches recently including human genomics, cancer, metagenomics, plant sciences, etc., moreover, it also has potential applications in the field of healthcare due to its fast turn-around time, portable and real-time data analysis. Those features make it a promising technology for the point-of-care testing (POCT) and its potential clinical applications are briefly discussed in this chapter.</p>', 'date' => '2020-01-01', 'pmid' => 'http://www.pubmed.gov/32710311', 'doi' => '10.1007/978-1-0716-0904-0_2,', 'modified' => '2020-09-01 14:39:13', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 4 => array( 'id' => '3045', 'name' => 'Condition-dependent co-regulation of genomic clusters of virulence factors in the grapevine trunk pathogen Neofusicoccum parvum', 'authors' => 'Massonnet M. et al.', 'description' => '<p>The ascomycete Neofusicoccum parvum, one of the causal agents of Botryosphaeria dieback, is a destructive wood-infecting fungus and a serious threat to grape production worldwide. The capability of colonizing woody tissue combined with the secretion of phytotoxic compounds is thought to underlie its pathogenicity and virulence. Here, we describe the repertoire of virulence factors and their transcriptional dynamics as the fungus feeds on different substrates and colonizes the woody stem. We assembled and annotated a highly contiguous genome using single molecule real-time DNA sequencing. Transcriptome profiling by RNA-sequencing determined the genome-wide patterns of expression of virulence factors both in vitro (potato dextrose agar or medium amended with grape wood as substrate) and in planta. Pairwise statistical testing of differential expression followed by co-expression network analysis revealed that physically clustered genes coding for putative virulence functions were induced depending on substrate or stage of plant infection. Co-expressed gene clusters were significantly enriched not only in genes associated with secondary metabolism, but also with cell wall degradation, suggesting that dynamic co-regulation of transcriptional networks contribute to multiple aspects of N. parvum virulence. In most of the co-expressed clusters, all genes shared at least a common motif in their promoter region indicative of co-regulation by the same transcription factor. Co-expression analysis also identified chromatin regulators with correlated expression with inducible clusters of virulence factors, suggesting a complex, multi-layered regulation of the virulence repertoire of N. parvum.</p>', 'date' => '2016-09-08', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27608421', 'doi' => '', 'modified' => '2016-10-10 10:53:20', 'created' => '2016-10-10 10:53:20', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 5 => array( 'id' => '2839', 'name' => 'Complete genome of Staphylococcus aureus Tager 104 provides evidence of its relation to modern systemic hospital-acquired strains', 'authors' => 'Richard W. DavisIV, Andrew D. Brannen, Mohammad J. Hossain, Scott Monsma, Paul E. Bock, Matthias Nahrendorf, David Mead, Michael Lodes, Mark R. Liles and Peter Panizzi', 'description' => '<section xmlns="" xmlns:meta="http://www.springer.com/app/meta" class="Abstract" id="Abs1" lang="en"> <div class="js-CollapseSection"> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec1"> <h5 class="Heading">Background</h5> <p id="Par1" class="Para"><em xmlns="" class="EmphasisTypeItalic">Staphylococcus aureus</em> (<em xmlns="" class="EmphasisTypeItalic">S. aureus</em>) infections range in severity due to expression of certain virulence factors encoded on mobile genetic elements (MGE). As such, characterization of these MGE, as well as single nucleotide polymorphisms, is of high clinical and microbiological importance. To understand the evolution of these dangerous pathogens, it is paramount to define reference strains that may predate MGE acquisition. One such candidate is <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> Tager 104, a previously uncharacterized strain isolated from a patient with impetigo in 1947.</p> </div> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec2"> <h5 class="Heading">Results</h5> <p id="Par2" class="Para">We show here that <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> Tager 104 can survive in the bloodstream and infect naïve organs. We also demonstrate a procedure to construct and validate the assembly of <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> genomes, using Tager 104 as a proof-of-concept. In so doing, we bridged confounding gap regions that limited our initial attempts to close this 2.82 Mb genome, through integration of data from Illumina Nextera paired-end, PacBio RS, and Lucigen NxSeq mate-pair libraries. Furthermore, we provide independent confirmation of our segmental arrangement of the Tager 104 genome by the sole use of Lucigen NxSeq libraries filled by paired-end MiSeq reads and alignment with SPAdes software. Genomic analysis of Tager 104 revealed limited MGE, and a νSaβ island configuration that is reminiscent of other hospital acquired <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> genomes.</p> </div> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec3"> <h5 class="Heading">Conclusions</h5> <p id="Par3" class="Para">Tager 104 represents an early-branching ancestor of certain hospital-acquired strains. Combined with its earlier isolation date and limited content of MGE, Tager 104 can serve as a viable reference for future comparative genome studies.</p> </div> </div> </section>', 'date' => '2016-03-03', 'pmid' => 'http://pubmed.gov/26940863', 'doi' => '10.1186/s12864-016-2433-8', 'modified' => '2016-03-09 10:13:09', 'created' => '2016-03-09 10:13:09', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 6 => array( 'id' => '2953', 'name' => 'Complete Genome Sequences of Four Escherichia coli ST95 Isolates from Bloodstream Infections', 'authors' => 'Craig M. Stephens, Jeffrey M. Skerker, Manraj S. Sekhon, Adam P. Arkin, Lee W. Rileyd', 'description' => '<p>Finished genome sequences are presented for four Escherichia coli strains isolated from bloodstream infections at San Francisco General Hospital. These strains provide reference sequences for four major fimH-identified sublineages within the multilocus sequence type (MLST) ST95 group, and provide insights into pathogenicity and differential antimicrobial susceptibility within this group.</p>', 'date' => '2015-11-05', 'pmid' => 'http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4645194/', 'doi' => '10.1128/genomeA.01241-15', 'modified' => '2016-06-13 15:50:25', 'created' => '2016-06-13 15:50:25', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 7 => array( 'id' => '2372', 'name' => 'Diversification of bacterial genome content through distinct mechanisms over different timescales.', 'authors' => 'Croucher NJ, Coupland PG, Stevenson AE, Callendrello A, Bentley SD, Hanage WP', 'description' => 'Bacterial populations often consist of multiple co-circulating lineages. Determining how such population structures arise requires understanding what drives bacterial diversification. Using 616 systematically sampled genomes, we show that Streptococcus pneumoniae lineages are typically characterized by combinations of infrequently transferred stable genomic islands: those moving primarily through transformation, along with integrative and conjugative elements and phage-related chromosomal islands. The only lineage containing extensive unique sequence corresponds to a set of atypical unencapsulated isolates that may represent a distinct species. However, prophage content is highly variable even within lineages, suggesting frequent horizontal transmission that would necessitate rapidly diversifying anti-phage mechanisms to prevent these viruses sweeping through populations. Correspondingly, two loci encoding Type I restriction-modification systems able to change their specificity over short timescales through intragenomic recombination are ubiquitous across the collection. Hence short-term pneumococcal variation is characterized by movement of phage and intragenomic rearrangements, with the slower transfer of stable loci distinguishing lineages.', 'date' => '2014-11-19', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25407023', 'doi' => '', 'modified' => '2015-07-24 15:39:04', 'created' => '2015-07-24 15:39:04', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 8 => array( 'id' => '2838', 'name' => 'Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance', 'authors' => 'Mark R Davies, Matthew T Holden, Paul Coupland, Jonathan H K Chen, Carola Venturini, Timothy C Barnett, Nouri L Ben Zakour, Herman Tse, Gordon Dougan, Kwok-Yung Yuen & Mark J Walker', 'description' => '<p><span>A scarlet fever outbreak began in mainland China and Hong Kong in 2011 (refs. </span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref1" title="Chen, M. et al. Outbreak of scarlet fever associated with emm12 type group A Streptococcus in 2011 in Shanghai, China. Pediatr. Infect. Dis. J. 31, e158-e162 (2012)." id="ref-link-1">1</a><span>–</span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref6" title="Yang, P. et al. Characteristics of group A Streptococcus strains circulating during scarlet fever epidemic, Beijing, China, 2011. Emerg. Infect. Dis. 19, 909-915 (2013)." id="ref-link-2">6</a><span>). Macrolide- and tetracycline-resistant </span><i>Streptococcus pyogenes emm</i><span>12 isolates represent the majority of clinical cases. Recently, we identified two mobile genetic elements that were closely associated with </span><i>emm</i><span>12 outbreak isolates: the integrative and conjugative element ICE-</span><i>emm</i><span>12, encoding genes for tetracycline and macrolide resistance, and prophage </span><span class="mb">Φ</span><span>HKU.vir, encoding the superantigens SSA and SpeC, as well as the DNase Spd1 (ref. </span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref4" title="Tse, H. et al. Molecular characterization of the 2011 Hong Kong scarlet fever outbreak. J. Infect. Dis. 206, 341-351 (2012)." id="ref-link-3">4</a><span>). Here we sequenced the genomes of 141 </span><i>emm</i><span>12 isolates, including 132 isolated in Hong Kong between 2005 and 2011. We found that the introduction of several ICE-</span><i>emm</i><span>12 variants, </span><span class="mb">Φ</span><span>HKU.vir and a new prophage, </span><span class="mb">Φ</span><span>HKU.ssa, occurred in three distinct </span><i>emm</i><span>12 lineages late in the twentieth century. Acquisition of </span><i>ssa</i><span> and transposable elements encoding multidrug resistance genes triggered the expansion of scarlet fever–associated </span><i>emm</i><span>12 lineages in Hong Kong. The occurrence of multidrug-resistant </span><i>ssa</i><span>-harboring scarlet fever strains should prompt heightened surveillance within China and abroad for the dissemination of these mobile genetic elements.</span></p>', 'date' => '2014-11-17', 'pmid' => 'http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html', 'doi' => '10.1038/ng.3147', 'modified' => '2016-03-08 14:33:49', 'created' => '2016-03-08 14:33:21', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 9 => array( 'id' => '2262', 'name' => 'Microevolution of Burkholderia pseudomallei during an acute infection.', 'authors' => 'Limmathurotsakul D, Holden MT, Coupland P, Price EP, Chantratita N, Wuthiekanun V, Amornchai P, Parkhill J, Peacock SJ', 'description' => 'We used whole-genome sequencing to evaluate 69 independent colonies of Burkholderia pseudomallei isolated from seven body sites of a patient with acute disseminated melioidosis. Fourteen closely related genotypes were found, providing evidence for the rapid in vivo diversification of B. pseudomallei after inoculation and systemic spread.', 'date' => '2014-09-01', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24966357', 'doi' => '', 'modified' => '2015-07-24 15:39:03', 'created' => '2015-07-24 15:39:03', 'ProductsPublication' => array( [maximum depth reached] ) ) ), 'Testimonial' => array( (int) 0 => array( 'id' => '57', 'name' => 'VIB - Megaruptor', 'description' => '<p>At the VIB Department of Molecular Genetics we strive to implement the best technologies in our workflows. Recent advances in long read sequencing using Oxford Nanopore technology have made the investigation of complex genomic variation in the human genome possible. A crucial step in the beginning of the library preparation is the shearing of genomic DNA, and this is where we implement the Megaruptor. The straightforward protocol is efficient, requires minimal sample handling and results in very reproducible fragment sizes. Another advantage is the consumable cost, which is lower than alternative technologies. For our applications we prefer shearing to lengths of 10 to 40kb, and for this the Megaruptor is the optimal solution.</p>', 'author' => 'Wouter de Coster and the Genomic Service Facility, VIB Department of Molecular Genetics, Antwerp, Belgium', 'featured' => true, 'slug' => 'vib-antwerp', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:35:57', 'created' => '2016-07-06 10:35:57', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '42', 'name' => 'Norwegian Sequencing Center - Megaruptor', 'description' => '<div class="page" title="Page 1"> <div class="layoutArea"> <div class="column"> <p><span>The Norwegian sequencing centre (NSC) is a national core facility fully equipped to handle a diverse set of sequencing projects. We see an increased interest in long read technologies, in which our PacBio Single Molecule sequencing service is important. PacBio, (or SMRT) sequencing, like any other single molecule system, starts with a critical library preparation step to generate long fragment libraries of 10-20kb length. Even longer libraries can be made, but common to all library types is to start with good quality DNA of high molecular weight. Physical shearing of the DNA, targeting the desired library length, is the first critical step in the procedure. NSC has experience with different methods to fragment DNA like nebulization, enzymatic, tagmentation, covaris AFA, g-tubes (also covaris), custom syringe/needle protocols and hydroshearing. The best fragmentation method should produce a sharp fragmentation pattern close to the target length as defined by the protocol settings, wi</span><span>th as little “smear like” pattern of lower molecular length as possible. </span><span>The final step of PacBio library preparation is to size select only the longest fragments (> 7kb) as this increases the overall success rate of only sequencing the longest library fragments. It is not uncommon to loose a lot of material during this step. However, if the initial fragmentation yields sharp fragmentation patterns with minimal smear, more of the total DNA is retained in the final library. This is perhaps most important on samples were DNA amounts are limited. The optimal fragmentation method has to produce similar fragmentation pattern each time, with little deviation with sample type and input amount. We have found the new Megaruptor from Diagenode to be one of the best instruments for routine shearing of DNA to lengths of 2-40kb. It is easy to use, with walk away shearing, and the instrument does all the washes before, in between samples and after shearing without user intervention. Disposable hydropores is beneficial for lowering the contamination risk. The fact that it is so easy to use make it a good choice for laboratories with multiple users and/or when training new staff. </span></p> </div> </div> </div>', 'author' => 'Morten Skage and Dr Ave Tooming-Klunderud, Norwegian Sequencing Centre, Oslo, Norway', 'featured' => false, 'slug' => 'norwegian-sequencing-center-megaruptor', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-03-22 11:31:43', 'created' => '2015-11-26 09:21:28', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '39', 'name' => 'Megaruptor® ', 'description' => '<p><span>We will use the Megaruptor<sup>®</sup> within our PacBio<sup>®</sup> Single Molecule Sequencing workflows, especially for the construction of very big insert (>20kb) libraries.</span></p>', 'author' => 'Andrea Patrignani, Anna Bratus and Ralph Schlapbach, Functional Genomics Center Zurich (ETHZ/UZH), Switzerland', 'featured' => false, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-11-23 10:04:40', 'created' => '2015-11-23 10:03:27', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 3 => array( 'id' => '3', 'name' => 'Megaruptor - Sylke Winkler', 'description' => '<p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p>', 'author' => 'Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany', 'featured' => true, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:38:08', 'created' => '2014-10-28 23:47:30', 'ProductsTestimonial' => array( [maximum depth reached] ) ) ), 'Area' => array(), 'SafetySheet' => array() ) $country = 'US' $countries_allowed = array( (int) 0 => 'CA', (int) 1 => 'US', (int) 2 => 'IE', (int) 3 => 'GB', (int) 4 => 'DK', (int) 5 => 'NO', (int) 6 => 'SE', (int) 7 => 'FI', (int) 8 => 'NL', (int) 9 => 'BE', (int) 10 => 'LU', (int) 11 => 'FR', (int) 12 => 'DE', (int) 13 => 'CH', (int) 14 => 'AT', (int) 15 => 'ES', (int) 16 => 'IT', (int) 17 => 'PT' ) $outsource = false $other_formats = array() $edit = '' $testimonials = '<blockquote><div class="page" title="Page 1"> <div class="layoutArea"> <div class="column"> <p><span>The Norwegian sequencing centre (NSC) is a national core facility fully equipped to handle a diverse set of sequencing projects. We see an increased interest in long read technologies, in which our PacBio Single Molecule sequencing service is important. PacBio, (or SMRT) sequencing, like any other single molecule system, starts with a critical library preparation step to generate long fragment libraries of 10-20kb length. Even longer libraries can be made, but common to all library types is to start with good quality DNA of high molecular weight. Physical shearing of the DNA, targeting the desired library length, is the first critical step in the procedure. NSC has experience with different methods to fragment DNA like nebulization, enzymatic, tagmentation, covaris AFA, g-tubes (also covaris), custom syringe/needle protocols and hydroshearing. The best fragmentation method should produce a sharp fragmentation pattern close to the target length as defined by the protocol settings, wi</span><span>th as little “smear like” pattern of lower molecular length as possible. </span><span>The final step of PacBio library preparation is to size select only the longest fragments (> 7kb) as this increases the overall success rate of only sequencing the longest library fragments. It is not uncommon to loose a lot of material during this step. However, if the initial fragmentation yields sharp fragmentation patterns with minimal smear, more of the total DNA is retained in the final library. This is perhaps most important on samples were DNA amounts are limited. The optimal fragmentation method has to produce similar fragmentation pattern each time, with little deviation with sample type and input amount. We have found the new Megaruptor from Diagenode to be one of the best instruments for routine shearing of DNA to lengths of 2-40kb. It is easy to use, with walk away shearing, and the instrument does all the washes before, in between samples and after shearing without user intervention. Disposable hydropores is beneficial for lowering the contamination risk. The fact that it is so easy to use make it a good choice for laboratories with multiple users and/or when training new staff. </span></p> </div> </div> </div><cite>Morten Skage and Dr Ave Tooming-Klunderud, Norwegian Sequencing Centre, Oslo, Norway</cite></blockquote> <blockquote><p><span>We will use the Megaruptor<sup>®</sup> within our PacBio<sup>®</sup> Single Molecule Sequencing workflows, especially for the construction of very big insert (>20kb) libraries.</span></p><cite>Andrea Patrignani, Anna Bratus and Ralph Schlapbach, Functional Genomics Center Zurich (ETHZ/UZH), Switzerland</cite></blockquote> ' $featured_testimonials = '<blockquote><span class="label-green" style="margin-bottom:16px;margin-left:-22px">TESTIMONIAL</span><p>At the VIB Department of Molecular Genetics we strive to implement the best technologies in our workflows. Recent advances in long read sequencing using Oxford Nanopore technology have made the investigation of complex genomic variation in the human genome possible. A crucial step in the beginning of the library preparation is the shearing of genomic DNA, and this is where we implement the Megaruptor. The straightforward protocol is efficient, requires minimal sample handling and results in very reproducible fragment sizes. Another advantage is the consumable cost, which is lower than alternative technologies. For our applications we prefer shearing to lengths of 10 to 40kb, and for this the Megaruptor is the optimal solution.</p><cite>Wouter de Coster and the Genomic Service Facility, VIB Department of Molecular Genetics, Antwerp, Belgium</cite></blockquote> <blockquote><span class="label-green" style="margin-bottom:16px;margin-left:-22px">TESTIMONIAL</span><p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p><cite>Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany</cite></blockquote> ' $testimonial = array( 'id' => '3', 'name' => 'Megaruptor - Sylke Winkler', 'description' => '<p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p>', 'author' => 'Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany', 'featured' => true, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:38:08', 'created' => '2014-10-28 23:47:30', 'ProductsTestimonial' => array( 'id' => '24', 'product_id' => '1824', 'testimonial_id' => '3' ) ) $related_products = '<li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydro-tubes-50-pc"><img src="/img/product/shearing_technologies/C30010018%20_hydro_tubes.png" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">C30010018</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2647" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2647" id="CartAdd/2647Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2647" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydro Tubes</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydro Tubes', 'C30010018', '65', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydro Tubes', 'C30010018', '65', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydro-tubes-50-pc" data-reveal-id="cartModal-2647" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydro Tubes</h6> </div> </div> </li> <li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydropore-short-10-pc"><img src="/img/product/shearing_technologies/hydropores.jpg" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">E07010001</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2645" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2645" id="CartAdd/2645Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2645" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydropore - short</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - short', 'E07010001', '160', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - short', 'E07010001', '160', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydropore-short-10-pc" data-reveal-id="cartModal-2645" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydropore - short</h6> </div> </div> </li> <li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydropore-long-10-pc"><img src="/img/product/shearing_technologies/hydropores.jpg" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">E07010002</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2646" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2646" id="CartAdd/2646Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2646" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydropore - long</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - long', 'E07010002', '170', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - long', 'E07010002', '170', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydropore-long-10-pc" data-reveal-id="cartModal-2646" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydropore - long</h6> </div> </div> </li> ' $related = array( 'id' => '2646', 'antibody_id' => null, 'name' => 'Hydropore - long', 'description' => '<p><span style="font-weight: 400;">Hydropore-longは<strong>10 kb - 75 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010002', 'old_catalog_number' => '', 'sf_code' => 'E07010002-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '145', 'price_USD' => '170', 'price_GBP' => '130', 'price_JPY' => '22715', 'price_CNY' => '', 'price_AUD' => '425', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-long-10-pc', 'meta_title' => 'Hydropore - long', 'meta_keywords' => '', 'meta_description' => 'Hydropore - long', 'modified' => '2022-04-18 09:02:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( 'id' => '68', 'product_id' => '1824', 'related_id' => '2646' ), 'Image' => array( (int) 0 => array( 'id' => '113', 'name' => 'product/shearing_technologies/hydropores.jpg', 'alt' => 'some alt', 'modified' => '2015-06-10 17:28:55', 'created' => '2015-06-10 17:28:55', 'ProductsImage' => array( [maximum depth reached] ) ) ) ) $rrbs_service = array( (int) 0 => (int) 1894, (int) 1 => (int) 1895 ) $chipseq_service = array( (int) 0 => (int) 2683, (int) 1 => (int) 1835, (int) 2 => (int) 1836, (int) 3 => (int) 2684, (int) 4 => (int) 1838, (int) 5 => (int) 1839, (int) 6 => (int) 1856 ) $labelize = object(Closure) { } $old_catalog_number = '' $label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>' $document = array( 'id' => '783', 'name' => 'Megaruptor® DNA Shearing System User Manual v2', 'description' => '<div class="page" title="Page 4"> <div class="layoutArea"> <div class="column"> <p><span>The Megaruptor</span><span>® </span><span>was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA in the range of 2 kb - 75 kb. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p> </div> </div> </div>', 'image_id' => null, 'type' => 'Manual', 'url' => 'files/products/shearing_technology/megaruptor/Megaruptor_manual.pdf', 'slug' => 'megaruptor-manual', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-07-23 17:29:32', 'created' => '2015-07-07 11:47:45', 'ProductsDocument' => array( 'id' => '656', 'product_id' => '1824', 'document_id' => '783' ) ) $publication = array( 'id' => '2262', 'name' => 'Microevolution of Burkholderia pseudomallei during an acute infection.', 'authors' => 'Limmathurotsakul D, Holden MT, Coupland P, Price EP, Chantratita N, Wuthiekanun V, Amornchai P, Parkhill J, Peacock SJ', 'description' => 'We used whole-genome sequencing to evaluate 69 independent colonies of Burkholderia pseudomallei isolated from seven body sites of a patient with acute disseminated melioidosis. Fourteen closely related genotypes were found, providing evidence for the rapid in vivo diversification of B. pseudomallei after inoculation and systemic spread.', 'date' => '2014-09-01', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24966357', 'doi' => '', 'modified' => '2015-07-24 15:39:03', 'created' => '2015-07-24 15:39:03', 'ProductsPublication' => array( 'id' => '203', 'product_id' => '1824', 'publication_id' => '2262' ) ) $externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/24966357" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755 View::_evaluate() - CORE/Cake/View/View.php, line 971 View::_render() - CORE/Cake/View/View.php, line 933 View::render() - CORE/Cake/View/View.php, line 473 Controller::render() - CORE/Cake/Controller/Controller.php, line 963 ProductsController::slug() - APP/Controller/ProductsController.php, line 1052 ReflectionMethod::invokeArgs() - [internal], line ?? Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491 Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193 Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167 [main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: campaign_id [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL --><div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"><?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>$viewFile = '/home/website-server/www/app/View/Products/view.ctp' $dataForView = array( 'language' => 'en', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'meta_title' => 'Megaruptor', 'product' => array( 'Product' => array( 'id' => '1824', 'antibody_id' => null, 'name' => 'Megaruptor<sup>®</sup>', 'description' => '<p><span>The Megaruptor</span><sup>®</sup><span> was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA from </span><strong>2 kb - 75 kb</strong><span>. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p>', 'label1' => 'Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores', 'info1' => '<h6><img src="https://www.diagenode.com/img/product/shearing_technologies/megaruptor-fig1.png" alt="" style="display: block; margin-left: auto; margin-right: auto;" width="851" height="245" /></h6> <h6 style="text-align: center;">Validation using two different DNA sources and two different methods of analysis. <strong>A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Fragment Analyzer profiles of human genomic DNA (25 ng/μl; 200 μl/sample) sheared at different software settings of 2, 3, 5 and 8 kb. (Standard Sensitivity Large Fragment Analysis Kit ; Advanced Analytical Technologies, Inc. was used for separation and fragment sizing).</h6>', 'label2' => '', 'info2' => '', 'label3' => '', 'info3' => '', 'format' => '1 unit', 'catalog_number' => 'B06010001', 'old_catalog_number' => '', 'sf_code' => null, 'type' => 'ACC', 'search_order' => '04-undefined', 'price_EUR' => 'on request', 'price_USD' => 'on request', 'price_GBP' => 'on request', 'price_JPY' => 'on request', 'price_CNY' => 'on request', 'price_AUD' => 'on request', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => true, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'megaruptor-1-unit', 'meta_title' => 'Megaruptor', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'modified' => '2019-02-20 12:43:01', 'created' => '2015-06-29 14:08:20', 'locale' => 'eng' ), 'Antibody' => array( 'host' => '*****', 'id' => null, 'name' => null, 'description' => null, 'clonality' => null, 'isotype' => null, 'lot' => null, 'concentration' => null, 'reactivity' => null, 'type' => null, 'purity' => null, 'classification' => null, 'application_table' => null, 'storage_conditions' => null, 'storage_buffer' => null, 'precautions' => null, 'uniprot_acc' => null, 'slug' => null, 'meta_keywords' => null, 'meta_description' => null, 'modified' => null, 'created' => null, 'select_label' => null ), 'Slave' => array(), 'Group' => array(), 'Related' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ) ), 'Application' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ) ), 'Category' => array(), 'Document' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ) ), 'Feature' => array(), 'Image' => array( (int) 0 => array( [maximum depth reached] ) ), 'Promotion' => array(), 'Protocol' => array(), 'Publication' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ), (int) 3 => array( [maximum depth reached] ), (int) 4 => array( [maximum depth reached] ), (int) 5 => array( [maximum depth reached] ), (int) 6 => array( [maximum depth reached] ), (int) 7 => array( [maximum depth reached] ), (int) 8 => array( [maximum depth reached] ), (int) 9 => array( [maximum depth reached] ) ), 'Testimonial' => array( (int) 0 => array( [maximum depth reached] ), (int) 1 => array( [maximum depth reached] ), (int) 2 => array( [maximum depth reached] ), (int) 3 => array( [maximum depth reached] ) ), 'Area' => array(), 'SafetySheet' => array() ) ) $language = 'en' $meta_keywords = '' $meta_description = 'Megaruptor' $meta_title = 'Megaruptor' $product = array( 'Product' => array( 'id' => '1824', 'antibody_id' => null, 'name' => 'Megaruptor<sup>®</sup>', 'description' => '<p><span>The Megaruptor</span><sup>®</sup><span> was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA from </span><strong>2 kb - 75 kb</strong><span>. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p>', 'label1' => 'Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores', 'info1' => '<h6><img src="https://www.diagenode.com/img/product/shearing_technologies/megaruptor-fig1.png" alt="" style="display: block; margin-left: auto; margin-right: auto;" width="851" height="245" /></h6> <h6 style="text-align: center;">Validation using two different DNA sources and two different methods of analysis. <strong>A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Fragment Analyzer profiles of human genomic DNA (25 ng/μl; 200 μl/sample) sheared at different software settings of 2, 3, 5 and 8 kb. (Standard Sensitivity Large Fragment Analysis Kit ; Advanced Analytical Technologies, Inc. was used for separation and fragment sizing).</h6>', 'label2' => '', 'info2' => '', 'label3' => '', 'info3' => '', 'format' => '1 unit', 'catalog_number' => 'B06010001', 'old_catalog_number' => '', 'sf_code' => null, 'type' => 'ACC', 'search_order' => '04-undefined', 'price_EUR' => 'on request', 'price_USD' => 'on request', 'price_GBP' => 'on request', 'price_JPY' => 'on request', 'price_CNY' => 'on request', 'price_AUD' => 'on request', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => true, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'megaruptor-1-unit', 'meta_title' => 'Megaruptor', 'meta_keywords' => '', 'meta_description' => 'Megaruptor', 'modified' => '2019-02-20 12:43:01', 'created' => '2015-06-29 14:08:20', 'locale' => 'eng' ), 'Antibody' => array( 'host' => '*****', 'id' => null, 'name' => null, 'description' => null, 'clonality' => null, 'isotype' => null, 'lot' => null, 'concentration' => null, 'reactivity' => null, 'type' => null, 'purity' => null, 'classification' => null, 'application_table' => null, 'storage_conditions' => null, 'storage_buffer' => null, 'precautions' => null, 'uniprot_acc' => null, 'slug' => null, 'meta_keywords' => null, 'meta_description' => null, 'modified' => null, 'created' => null, 'select_label' => null ), 'Slave' => array(), 'Group' => array(), 'Related' => array( (int) 0 => array( 'id' => '2647', 'antibody_id' => null, 'name' => 'Hydro Tubes', 'description' => '<p><span>Hydro Tubes for Megaruptor</span><sup>®</sup><span> The Microcentrifuge Tubes (0.5 ml) have been thoroughly validated for use on the Megaruptor</span><sup>®</sup><span> (B06010001) and are strongly recommended for DNA shearing applications using this instrument.</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => 'Characteristics', 'info1' => '<ul> <li>Sterile</li> <li>Natural</li> <li>Autoclavable</li> <li>Freezable</li> </ul> <p></p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '50 pc', 'catalog_number' => 'C30010018', 'old_catalog_number' => '', 'sf_code' => 'C30010018-1860', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '65', 'price_USD' => '65', 'price_GBP' => '60', 'price_JPY' => '10180', 'price_CNY' => '', 'price_AUD' => '162', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydro-tubes-50-pc', 'meta_title' => 'Hydro Tubes', 'meta_keywords' => '', 'meta_description' => 'Hydro Tubes', 'modified' => '2020-04-07 05:19:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '2645', 'antibody_id' => null, 'name' => 'Hydropore - short', 'description' => '<p><span style="font-weight: 400;">Hydropore-shortは<strong>2 kb - 9 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010001', 'old_catalog_number' => '', 'sf_code' => 'E07010001-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '135', 'price_USD' => '160', 'price_GBP' => '125', 'price_JPY' => '21150', 'price_CNY' => '', 'price_AUD' => '400', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-short-10-pc', 'meta_title' => 'Hydropore - short', 'meta_keywords' => '', 'meta_description' => 'Hydropore - short', 'modified' => '2022-04-18 09:01:09', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '2646', 'antibody_id' => null, 'name' => 'Hydropore - long', 'description' => '<p><span style="font-weight: 400;">Hydropore-longは<strong>10 kb - 75 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010002', 'old_catalog_number' => '', 'sf_code' => 'E07010002-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '145', 'price_USD' => '170', 'price_GBP' => '130', 'price_JPY' => '22715', 'price_CNY' => '', 'price_AUD' => '425', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-long-10-pc', 'meta_title' => 'Hydropore - long', 'meta_keywords' => '', 'meta_description' => 'Hydropore - long', 'modified' => '2022-04-18 09:02:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( [maximum depth reached] ), 'Image' => array( [maximum depth reached] ) ) ), 'Application' => array( (int) 0 => array( 'id' => '13', 'position' => '10', 'parent_id' => '3', 'name' => 'DNA/RNA shearing', 'description' => '<div class="row"> <div class="small-12 medium-12 large-12 columns">In recent years, advances in Next-Generation Sequencing (NGS) have revolutionized genomics and biology. This growth has fueled demands on upstream techniques for optimal sample preparation and genomic library construction. One of the most critical aspects of optimal library preparation is the quality of the DNA to be sequenced. The DNA must first be effectively and consistently sheared into the appropriate fragment size (depending on the sequencing platform) to enable sensitive and reliable NGS results. The <strong>Bioruptor</strong><sup>®</sup> <strong>Pico</strong> and the <strong>Megaruptor</strong><sup>®</sup> provide superior sample yields, fragment size, and consistency, which are essential for Next-Generation Sequencing workflows. Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor<sup>®</sup></a>.</div> </div> <p></p> <div class="row"> <div class="small-7 medium-7 large-7 columns text-center"><img src="https://www.diagenode.com/img/applications/true-flexibility-with-br-ngs.jpg" /></div> <div class="small-5 medium-5 large-5 columns"><small><strong>Programmable DNA size distribution and high reproducibility with Bioruptor<sup>®</sup> Pico using 0.65 (panel A) or 0.1 ml (panel B) microtubes</strong>. <b>Panel A:</b> 200 bp after 13 cycles (13 sec ON/OFF) using 100 µl volume. Average size: 204; CV%:1.89%). <b>Panel B:</b> 200 bp after 20 cycles (30 sec ON/OFF) using 10 µl volume. (Average size: 215 bp; CV%: 6.6%). <b>Panel A & B:</b> peak electropherogram view. <b>Panel C & D:</b> virtual gel view.</small></div> </div> <p><br /><br /></p> <div class="row"> <div class="small-10 medium-10 large-10 columns text-center end small-offset-1"><img src="https://www.diagenode.com/img/applications/megaruptor-short-frag.jpg" /></div> <div class="small-12 medium-12 large-12 columns"><small><strong> Reproducible and narrow DNA size distribution with Megaruptor® using short fragment size Hydropores Validation using two different DNA sources and two different methods of analysis. A:</strong> Shearing of lambda phage genomic DNA (20 ng/μl; 150 μl/sample) sheared at different speed settings and analyzed on 1% agarose gel. <strong>B:</strong> Bioanalyzer profiles of human genomic DNA (20 ng/μl; 150 μl/sample) sheared at different software settings of 2 and 5 kb. Three independent experiments were run for each setting. (Agilent DNA 12000 kit was used for separation and fragment sizing).</small></div> </div> <p><br /><br /></p> <div class="row"> <div class="small-4 medium-4 large-4 columns text-center"><img src="https://www.diagenode.com/img/applications/megaruptor-long-frag.jpg" /></div> <div class="small-8 medium-8 large-8 columns"><small><strong> Demonstrated shearing to fragment sizes between 15 kb and 75 kb with Megaruptor® using long fragment size Hydropores. </strong>Image shows DNA size distribution of human genomic DNA sheared with long fragment Hydropores. DNA was analyzed by pulsed field gel electrophoresis (PFGE) in 1% agarose gel and a mean size of smears was estimated using Image Lab 4.1 software.<br /> * indicates unsheared DNA </small></div> </div>', 'in_footer' => false, 'in_menu' => true, 'online' => true, 'tabular' => false, 'slug' => 'dna-rna-shearing', 'meta_keywords' => 'DNA/RNA shearing,Bioruptor® Pico,Megaruptor®,Next-Generation Sequencing ', 'meta_description' => 'Bioruptor® Pico and the Megaruptor® provide superior sample yields, fragment size, and consistency, which are essential for Next-Generation Sequencing workflows.', 'meta_title' => 'DNA shearing & RNA shearing for Next-Generation Sequencing (NGS) | Diagenode', 'modified' => '2017-12-08 14:44:11', 'created' => '2014-10-29 12:45:41', 'ProductsApplication' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '3', 'position' => '10', 'parent_id' => null, 'name' => '次世代シーケンシング', 'description' => '<div class="row"> <div class="small-12 medium-12 large-12 columns"> <h2 style="font-size: 22px;">DNA断片化、ライブラリー調製、自動化:NGSのワンストップショップ</h2> <table class="small-12 medium-12 large-12 columns"> <tbody> <tr> <th class="small-12 medium-12 large-12 columns"> <h4>1. 断片化装置を選択してください:150 bp〜75 kbの範囲でDNAを断片化します。</h4> </th> </tr> <tr style="background-color: #ffffff;"> <td class="small-12 medium-12 large-12 columns"></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns"><a href="../p/bioruptor-pico-sonication-device"><img src="https://www.diagenode.com/img/product/shearing_technologies/bioruptor_pico.jpg" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/megaruptor2-1-unit"><img src="https://www.diagenode.com/img/product/shearing_technologies/B06010001_megaruptor2.jpg" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/bioruptor-one-sonication-device"><img src="https://www.diagenode.com/img/product/shearing_technologies/br-one-profil.png" /></a></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns">5μlまで断片化:150 bp〜2 kb<br />NGS DNAライブラリー調製およびFFPE核酸抽出に最適で、</td> <td class="small-4 medium-4 large-4 columns">2 kb〜75 kbの範囲をできます。<br />メイトペアライブラリー調製および長いフラグメントDNAシーケンシングに最適で、この軽量デスクトップデバイスで</td> <td class="small-4 medium-4 large-4 columns">20または50μlの断片化が可能です。</td> </tr> </tbody> </table> <table class="small-12 medium-12 large-12 columns"> <tbody> <tr> <th class="small-8 medium-8 large-8 columns"> <h4>2. 最適化されたライブラリー調整キットを選択してください。</h4> </th> <th class="small-4 medium-4 large-4 columns"> <h4>3. ライブラリー前処理自動化を選択して、比類のないデータ再現性を実感</h4> </th> </tr> <tr style="background-color: #ffffff;"> <td class="small-12 medium-12 large-12 columns"></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns"><a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns"><img src="https://www.diagenode.com/img/product/kits/microPlex_library_preparation.png" style="display: block; margin-left: auto; margin-right: auto;" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/ideal-library-preparation-kit-x24-incl-index-primer-set-1-24-rxns"><img src="https://www.diagenode.com/img/product/kits/box_kit.jpg" style="display: block; margin-left: auto; margin-right: auto;" height="173" width="250" /></a></td> <td class="small-4 medium-4 large-4 columns"><a href="../p/sx-8g-ip-star-compact-automated-system-1-unit"><img src="https://www.diagenode.com/img/product/automation/B03000002%20_ipstar_compact.png" style="display: block; margin-left: auto; margin-right: auto;" /></a></td> </tr> <tr> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">50pgの低入力:MicroPlex Library Preparation Kit</td> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">5ng以上:iDeal Library Preparation Kit</td> <td class="small-4 medium-4 large-4 columns" style="text-align: center;">Achieve great NGS data easily</td> </tr> </tbody> </table> </div> </div> <blockquote> <div class="row"> <div class="small-12 medium-12 large-12 columns"><span class="label" style="margin-bottom: 16px; margin-left: -22px; font-size: 15px;">DiagenodeがNGS研究にぴったりなプロバイダーである理由</span> <p>Diagenodeは15年以上もエピジェネティクス研究に専念、専門としています。 ChIP研究クロマチン用のユニークな断片化システムの開発から始まり、 専門知識を活かし、5μlのせん断体積まで可能で、NGS DNAライブラリーの調製に最適な最先端DNA断片化装置の開発にたどり着きました。 我々は以来、ChIP-seq、Methyl-seq、NGSライブラリー調製用キットを研究開発し、業界をリードする免疫沈降研究と同様に、ライブラリー調製を自動化および完結させる独自の自動化システムを開発にも成功しました。</p> <ul> <li>信頼されるせん断装置</li> <li>様々なインプットからのライブラリ作成キット</li> <li>独自の自動化デバイス</li> </ul> </div> </div> </blockquote> <div class="row"> <div class="small-12 columns"> <ul class="accordion" data-accordion=""> <li class="accordion-navigation"><a href="#panel1a">次世代シーケンシングへの理解とその専門知識</a> <div id="panel1a" class="content"> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p><strong>次世代シーケンシング (NGS)</strong> )は、著しいスケールとハイスループットでシーケンシングを行い、1日に数十億もの塩基生成を可能にします。 NGSのハイスループットは迅速でありながら正確で、再現性のあるデータセットを実現し、さらにシーケンシング費用を削減します。 NGSは、ゲノムシーケンシング、ゲノム再シーケンシング、デノボシーケンシング、トランスクリプトームシーケンシング、その他にDNA-タンパク質相互作用の検出やエピゲノムなどを示します。 指数関数的に増加するシーケンシングデータの需要は、計算分析の障害や解釈、データストレージなどの課題を解決します。</p> <p>アプリケーションおよび出発物質に応じて、数百万から数十億の鋳型DNA分子を大規模に並行してシーケンシングすることが可能です。その為に、異なる化学物質を使用するいくつかの市販のNGSプラットフォームを利用することができます。 NGSプラットフォームの種類によっては、事前準備とライブラリー作成が必要です。</p> <p>NGSにとっても、特にデータ処理と分析に関した大きな課題はあります。第3世代技術はゲノミクス研究にさらに革命を起こすであろうと大きく期待されています。</p> </div> </div> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <p><strong>NGS アプリケーション</strong></p> <ul> <li>全ゲノム配列決定</li> <li>デノボシーケンシング</li> <li>標的配列</li> <li>Exomeシーケンシング</li> <li>トランスクリプトーム配列決定</li> <li>ゲノム配列決定</li> <li>ミトコンドリア配列決定</li> <li>DNA-タンパク質相互作用(ChIP-seq</li> <li>バリアント検出</li> <li>ゲノム仕上げ</li> </ul> </div> <div class="small-6 medium-6 large-6 columns"> <p><strong>研究分野におけるNGS:</strong></p> <ul> <li>腫瘍学</li> <li>リプロダクティブ・ヘルス</li> <li>法医学ゲノミクス</li> <li>アグリゲノミックス</li> <li>複雑な病気</li> <li>微生物ゲノミクス</li> <li>食品・環境ゲノミクス</li> <li>創薬ゲノミクス - パーソナライズド・メディカル</li> </ul> </div> <div class="small-12 medium-12 large-12 columns"> <p><strong>NGSの用語</strong></p> <dl> <dt>リード(読み取り)</dt> <dd>この装置から得られた連続した単一のストレッチ</dd> <dt>断片リード</dt> <dd>フラグメントライブラリからの読み込み。 シーケンシングプラットフォームに応じて、読み取りは通常約100〜300bp。</dd> <dt>断片ペアエンドリード</dt> <dd>断片ライブラリーからDNA断片の各末端2つの読み取り。</dd> <dt>メイトペアリード</dt> <dd>大きなDNA断片(通常は予め定義されたサイズ範囲)の各末端から2つの読み取り。</dd> <dt>カバレッジ(例)</dt> <dd>30×適用範囲とは、参照ゲノム中の各塩基対が平均30回の読み取りを示す。</dd> </dl> </div> </div> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <h2>NGSプラットフォーム</h2> <h3><a href="http://www.illumina.com" target="_blank">イルミナ</a></h3> <p>イルミナは、クローン的に増幅された鋳型DNA(クラスター)上に位置する、蛍光標識された可逆的鎖ターミネーターヌクレオチドを用いた配列別合成技術を使用。 DNAクラスターは、ガラスフローセルの表面上に固定化され、 ワークフローは、4つのヌクレオチド(それぞれ異なる蛍光色素で標識された)の組み込み、4色イメージング、色素や末端基の切断、取り込み、イメージングなどを繰り返します。フローセルは大規模な並列配列決定を受ける。 この方法により、単一蛍光標識されたヌクレオチドの制御添加によるモノヌクレオチドのエラーを回避する可能性があります。 読み取りの長さは、通常約100〜150 bpです。</p> <h3><a href="http://www.lifetechnologies.com" target="_blank">イオン トレント</a></h3> <p>イオントレントは、半導体技術チップを用いて、合成中にヌクレオチドを取り込む際に放出されたプロトンを検出します。 これは、イオン球粒子と呼ばれるビーズの表面にエマルションPCR(emPCR)を使用し、リンクされた特定のアダプターを用いてDNA断片を増幅します。 各ビーズは1種類のDNA断片で覆われていて、異なるDNA断片を有するビーズは次いで、チップの陽子感知ウェル内に配置されます。 チップには一度に4つのヌクレオチドのうちの1つが浸水し、このプロセスは異なるヌクレオチドで15秒ごとに繰り返されます。 配列決定の間に4つの塩基の各々が1つずつ導入されます、組み込みの場合はプロトンが放出され、電圧信号が取り込みに比例して検出されます。.</p> <h3><a href="http://www.pacificbiosciences.com" target="_blank">パシフィック バイオサイエンス</a></h3> <p>パシフィックバイオサイエンスでは、20kbを超える塩基対の読み取りも、単一分子リアルタイム(SMRT)シーケンシングによる構造および細胞タイプの変化を観察することができます。 このプラットフォームでは、超長鎖二本鎖DNA(dsDNA)断片が、Megaruptor(登録商標)のようなDiagenode装置を用いたランダムシアリングまたは目的の標的領域の増幅によって生成されます。 SMRTbellライブラリーは、ユニバーサルヘアピンアダプターをDNA断片の各末端に連結することによって生成します。 サイズ選択条件による洗浄ステップの後、配列決定プライマーをSMRTbellテンプレートにアニーリングし、鋳型DNAに結合したDNAポリメラーゼを含む配列決定を、蛍光標識ヌクレオチドの存在下で開始。 各塩基が取り込まれると、異なる蛍光のパルスをリアルタイムで検出します。</p> <h3><a href="https://nanoporetech.com" target="_blank">オックスフォード ナノポア</a></h3> <p>Oxford Nanoporeは、単一のDNA分子配列決定に基づく技術を開発します。その技術により生物学的分子、すなわちDNAが一群の電気抵抗性高分子膜として位置するナノスケールの孔(ナノ細孔)またはその近くを通過し、イオン電流が変化します。 この変化に関する情報は、例えば4つのヌクレオチド(AまたはG r CまたはT)ならびに修飾されたヌクレオチドすべてを区別することによって分子情報に訳されます。 シーケンシングミニオンデバイスのフローセルは、数百個のナノポアチャネルのセンサアレイを含みます。 DNAサンプルは、Diagenode社のMegaruptor(登録商標)を用いてランダムシアリングによって生成され得る超長鎖DNAフラグメントが必要です。</p> <h3><a href="http://www.lifetechnologies.com/be/en/home/life-science/sequencing/next-generation-sequencing/solid-next-generation-sequencing.html" target="_blank">SOLiD</a></h3> <p>SOLiDは、ユニークな化学作用により、何千という個々のDNA分子の同時配列決定を可能にします。 それは、アダプター対ライブラリーのフラグメントが適切で、せん断されたゲノムDNAへのアダプターのライゲーションによるライブラリー作製から始まります。 次のステップでは、エマルジョンPCR(emPCR)を実施して、ビーズの表面上の個々の鋳型DNA分子をクローン的に増幅。 emPCRでは、個々の鋳型DNAをPCR試薬と混合し、水中油型エマルジョン内の疎水性シェルで囲まれた水性液滴内のプライマーコートビーズを、配列決定のためにロードするスライドガラスの表面にランダムに付着。 この技術は、シークエンシングプライマーへのライゲーションで競合する4つの蛍光標識されたジ塩基プローブのセットを使用します。</p> <h3><a href="http://454.com/products/technology.asp" target="_blank">454</a></h3> <p>454は、大規模並列パイロシーケンシングを利用しています。 始めに全ゲノムDNAまたは標的遺伝子断片の300〜800bp断片のライブラリー調製します。 次に、DNAフラグメントへのアダプターの付着および単一のDNA鎖の分離。 その後アダプターに連結されたDNAフラグメントをエマルジョンベースのクローン増幅(emPCR)で処理し、DNAライブラリーフラグメントをミクロンサイズのビーズ上に配置します。 各DNA結合ビーズを光ファイバーチップ上のウェルに入れ、器具に挿入します。 4つのDNAヌクレオチドは、配列決定操作中に固定された順序で連続して加えられ、並行して配列決定されます。</p> </div> </div> </div> </li> </ul> </div> </div>', 'in_footer' => true, 'in_menu' => true, 'online' => true, 'tabular' => true, 'slug' => 'next-generation-sequencing', 'meta_keywords' => 'Next-generation sequencing,NGS,Whole genome sequencing,NGS platforms,DNA/RNA shearing', 'meta_description' => 'Diagenode offers kits and DNA/RNA shearing technology prior to next-generation sequencing, many Next-generation sequencing platforms require preparation of the DNA.', 'meta_title' => 'Next-generation sequencing (NGS) Applications and Methods | Diagenode', 'modified' => '2018-07-26 05:24:29', 'created' => '2015-04-01 22:47:04', 'ProductsApplication' => array( [maximum depth reached] ) ) ), 'Category' => array(), 'Document' => array( (int) 0 => array( 'id' => '14', 'name' => 'Improvements in Hydrodynamic Shearing Facilitates 20-50 kb True Mate Pair Library Construction', 'description' => '<p><span>The Megaruptor<sup>®</sup> has been used by several sequencing centers to produce high-quality small insert fragment libraries for Illumina and PacBio platforms. David Mead and colleagues from Lucigen<sup>®</sup> have used the Megaruptor<sup>®</sup> to prepare 20-50 kb inserts for a new mate-pair library technology that is >70% efficient. </span></p>', 'image_id' => null, 'type' => 'Poster', 'url' => 'files/posters/Mead_et_al_Lucigen_Diagenode_Poster.pdf', 'slug' => 'mead-et-al-lucigen-diagenode-poster', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-09-29 13:42:27', 'created' => '2015-07-03 16:05:15', 'ProductsDocument' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '783', 'name' => 'Megaruptor® DNA Shearing System User Manual v2', 'description' => '<div class="page" title="Page 4"> <div class="layoutArea"> <div class="column"> <p><span>The Megaruptor</span><span>® </span><span>was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA in the range of 2 kb - 75 kb. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p> </div> </div> </div>', 'image_id' => null, 'type' => 'Manual', 'url' => 'files/products/shearing_technology/megaruptor/Megaruptor_manual.pdf', 'slug' => 'megaruptor-manual', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-07-23 17:29:32', 'created' => '2015-07-07 11:47:45', 'ProductsDocument' => array( [maximum depth reached] ) ) ), 'Feature' => array(), 'Image' => array( (int) 0 => array( 'id' => '139', 'name' => 'product/shearing_technologies/B06010001_megaruptor.jpg', 'alt' => 'megaruptor', 'modified' => '2015-07-23 17:39:35', 'created' => '2015-06-10 17:28:55', 'ProductsImage' => array( [maximum depth reached] ) ) ), 'Promotion' => array(), 'Protocol' => array(), 'Publication' => array( (int) 0 => array( 'id' => '4018', 'name' => 'Complete Genome Sequence of the Type Strain Pectobacterium punjabenseSS95, Isolated from a Potato Plant with Blackleg Symptoms.', 'authors' => 'Sarfraz, S and Oulghazi, S and Cigna, J and Sahi, ST and Riaz, K andTufail, MR and Fayyaz, A and Naveed, K and Hameed, A and Lopez-Roques, Cand Vandecasteele, C and Faure, D', 'description' => '<p>Pectobacterium punjabense is a newly described species causing blackleg disease in potato plants. Therefore, by the combination of long (Oxford Nanopore Technologies, MinION) and short (Illumina MiSeq) reads, we sequenced the complete genome of P. punjabense SS95T, which contains a circular chromosome of 4.793 Mb with a GC content of 50.7\%.</p>', 'date' => '2020-08-06', 'pmid' => 'http://www.pubmed.gov/32763925', 'doi' => '10.1128/MRA.00420-20.', 'modified' => '2020-12-16 17:37:51', 'created' => '2020-10-12 14:54:59', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '4001', 'name' => 'Sizing, stabilising, and cloning repeat-expansions for gene targeting constructs.', 'authors' => 'Nair RR, Tibbit C, Thompson D, McLeod R, Nakhuda A, Simon MM, Baloh RH, Fisher EMC, Isaacs AM, Cunningham TJ', 'description' => '<p>Aberrant microsatellite repeat-expansions at specific loci within the human genome cause several distinct, heritable, and predominantly neurological, disorders. Creating models for these diseases poses a challenge, due to the instability of such repeats in bacterial vectors, especially with large repeat expansions. Designing constructs for more precise genome engineering projects, such as engineering knock-in mice, proves a greater challenge still, since these unstable repeats require numerous cloning steps in order to introduce homology arms or selection cassettes. Here, we report our efforts to clone a large hexanucleotide repeat in the C9orf72 gene, originating from within a BAC construct, derived from a C9orf72-ALS patient. We provide detailed methods for efficient repeat sizing and growth conditions in bacteria to facilitate repeat retention during growth and sub-culturing. We report that sub-cloning into a linear vector dramatically improves stability, but is dependent on the relative orientation of DNA replication through the repeat, consistent with previous studies. We envisage the findings presented here provide a relatively straightforward route to maintaining large-range microsatellite repeat-expansions, for efficient cloning into vectors.</p>', 'date' => '2020-07-25', 'pmid' => 'http://www.pubmed.gov/32721467', 'doi' => '10.1016/j.ymeth.2020.07.007', 'modified' => '2020-09-01 14:39:41', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '3980', 'name' => 'Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants.', 'authors' => 'Xu Z, Pu X, Gao R, Demurtas OC, Fleck SJ, Richter M, He C, Ji A, Sun W, Kong J, Hu K, Ren F, Song J, Wang Z, Gao T, Xiong C, Yu H, Xin T, Albert VA, Giuliano G, Chen S, Song J', 'description' => '<p>BACKGROUND: Plants have evolved a panoply of specialized metabolites that increase their environmental fitness. Two examples are caffeine, a purine psychotropic alkaloid, and crocins, a group of glycosylated apocarotenoid pigments. Both classes of compounds are found in a handful of distantly related plant genera (Coffea, Camellia, Paullinia, and Ilex for caffeine; Crocus, Buddleja, and Gardenia for crocins) wherein they presumably evolved through convergent evolution. The closely related Coffea and Gardenia genera belong to the Rubiaceae family and synthesize, respectively, caffeine and crocins in their fruits. RESULTS: Here, we report a chromosomal-level genome assembly of Gardenia jasminoides, a crocin-producing species, obtained using Oxford Nanopore sequencing and Hi-C technology. Through genomic and functional assays, we completely deciphered for the first time in any plant the dedicated pathway of crocin biosynthesis. Through comparative analyses with Coffea canephora and other eudicot genomes, we show that Coffea caffeine synthases and the first dedicated gene in the Gardenia crocin pathway, GjCCD4a, evolved through recent tandem gene duplications in the two different genera, respectively. In contrast, genes encoding later steps of the Gardenia crocin pathway, ALDH and UGT, evolved through more ancient gene duplications and were presumably recruited into the crocin biosynthetic pathway only after the evolution of the GjCCD4a gene. CONCLUSIONS: This study shows duplication-based divergent evolution within the coffee family (Rubiaceae) of two characteristic secondary metabolic pathways, caffeine and crocin biosynthesis, from a common ancestor that possessed neither complete pathway. These findings provide significant insights on the role of tandem duplications in the evolution of plant specialized metabolism.</p>', 'date' => '2020-06-18', 'pmid' => 'http://www.pubmed.gov/32552824', 'doi' => '10.1186/s12915-020-00795-3', 'modified' => '2020-09-01 15:27:14', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 3 => array( 'id' => '4002', 'name' => 'Nanopore Sequencing and Its Clinical Applications.', 'authors' => 'Sun X, Song L, Yang W, Zhang L, Liu M, Li X, Tian G, Wang W', 'description' => '<p>Nanopore sequencing is a method for determining the order and modifications of DNA/RNA nucleotides by detecting the electric current variations when DNA/RNA oligonucleotides pass through the nanometer-sized hole (nanopore). Nanopore-based DNA analysis techniques have been commercialized by Oxford Nanopore Technologies, NabSys, and Sequenom, and widely used in scientific researches recently including human genomics, cancer, metagenomics, plant sciences, etc., moreover, it also has potential applications in the field of healthcare due to its fast turn-around time, portable and real-time data analysis. Those features make it a promising technology for the point-of-care testing (POCT) and its potential clinical applications are briefly discussed in this chapter.</p>', 'date' => '2020-01-01', 'pmid' => 'http://www.pubmed.gov/32710311', 'doi' => '10.1007/978-1-0716-0904-0_2,', 'modified' => '2020-09-01 14:39:13', 'created' => '2020-08-21 16:41:39', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 4 => array( 'id' => '3045', 'name' => 'Condition-dependent co-regulation of genomic clusters of virulence factors in the grapevine trunk pathogen Neofusicoccum parvum', 'authors' => 'Massonnet M. et al.', 'description' => '<p>The ascomycete Neofusicoccum parvum, one of the causal agents of Botryosphaeria dieback, is a destructive wood-infecting fungus and a serious threat to grape production worldwide. The capability of colonizing woody tissue combined with the secretion of phytotoxic compounds is thought to underlie its pathogenicity and virulence. Here, we describe the repertoire of virulence factors and their transcriptional dynamics as the fungus feeds on different substrates and colonizes the woody stem. We assembled and annotated a highly contiguous genome using single molecule real-time DNA sequencing. Transcriptome profiling by RNA-sequencing determined the genome-wide patterns of expression of virulence factors both in vitro (potato dextrose agar or medium amended with grape wood as substrate) and in planta. Pairwise statistical testing of differential expression followed by co-expression network analysis revealed that physically clustered genes coding for putative virulence functions were induced depending on substrate or stage of plant infection. Co-expressed gene clusters were significantly enriched not only in genes associated with secondary metabolism, but also with cell wall degradation, suggesting that dynamic co-regulation of transcriptional networks contribute to multiple aspects of N. parvum virulence. In most of the co-expressed clusters, all genes shared at least a common motif in their promoter region indicative of co-regulation by the same transcription factor. Co-expression analysis also identified chromatin regulators with correlated expression with inducible clusters of virulence factors, suggesting a complex, multi-layered regulation of the virulence repertoire of N. parvum.</p>', 'date' => '2016-09-08', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27608421', 'doi' => '', 'modified' => '2016-10-10 10:53:20', 'created' => '2016-10-10 10:53:20', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 5 => array( 'id' => '2839', 'name' => 'Complete genome of Staphylococcus aureus Tager 104 provides evidence of its relation to modern systemic hospital-acquired strains', 'authors' => 'Richard W. DavisIV, Andrew D. Brannen, Mohammad J. Hossain, Scott Monsma, Paul E. Bock, Matthias Nahrendorf, David Mead, Michael Lodes, Mark R. Liles and Peter Panizzi', 'description' => '<section xmlns="" xmlns:meta="http://www.springer.com/app/meta" class="Abstract" id="Abs1" lang="en"> <div class="js-CollapseSection"> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec1"> <h5 class="Heading">Background</h5> <p id="Par1" class="Para"><em xmlns="" class="EmphasisTypeItalic">Staphylococcus aureus</em> (<em xmlns="" class="EmphasisTypeItalic">S. aureus</em>) infections range in severity due to expression of certain virulence factors encoded on mobile genetic elements (MGE). As such, characterization of these MGE, as well as single nucleotide polymorphisms, is of high clinical and microbiological importance. To understand the evolution of these dangerous pathogens, it is paramount to define reference strains that may predate MGE acquisition. One such candidate is <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> Tager 104, a previously uncharacterized strain isolated from a patient with impetigo in 1947.</p> </div> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec2"> <h5 class="Heading">Results</h5> <p id="Par2" class="Para">We show here that <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> Tager 104 can survive in the bloodstream and infect naïve organs. We also demonstrate a procedure to construct and validate the assembly of <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> genomes, using Tager 104 as a proof-of-concept. In so doing, we bridged confounding gap regions that limited our initial attempts to close this 2.82 Mb genome, through integration of data from Illumina Nextera paired-end, PacBio RS, and Lucigen NxSeq mate-pair libraries. Furthermore, we provide independent confirmation of our segmental arrangement of the Tager 104 genome by the sole use of Lucigen NxSeq libraries filled by paired-end MiSeq reads and alignment with SPAdes software. Genomic analysis of Tager 104 revealed limited MGE, and a νSaβ island configuration that is reminiscent of other hospital acquired <em xmlns="" class="EmphasisTypeItalic">S. aureus</em> genomes.</p> </div> <div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec3"> <h5 class="Heading">Conclusions</h5> <p id="Par3" class="Para">Tager 104 represents an early-branching ancestor of certain hospital-acquired strains. Combined with its earlier isolation date and limited content of MGE, Tager 104 can serve as a viable reference for future comparative genome studies.</p> </div> </div> </section>', 'date' => '2016-03-03', 'pmid' => 'http://pubmed.gov/26940863', 'doi' => '10.1186/s12864-016-2433-8', 'modified' => '2016-03-09 10:13:09', 'created' => '2016-03-09 10:13:09', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 6 => array( 'id' => '2953', 'name' => 'Complete Genome Sequences of Four Escherichia coli ST95 Isolates from Bloodstream Infections', 'authors' => 'Craig M. Stephens, Jeffrey M. Skerker, Manraj S. Sekhon, Adam P. Arkin, Lee W. Rileyd', 'description' => '<p>Finished genome sequences are presented for four Escherichia coli strains isolated from bloodstream infections at San Francisco General Hospital. These strains provide reference sequences for four major fimH-identified sublineages within the multilocus sequence type (MLST) ST95 group, and provide insights into pathogenicity and differential antimicrobial susceptibility within this group.</p>', 'date' => '2015-11-05', 'pmid' => 'http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4645194/', 'doi' => '10.1128/genomeA.01241-15', 'modified' => '2016-06-13 15:50:25', 'created' => '2016-06-13 15:50:25', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 7 => array( 'id' => '2372', 'name' => 'Diversification of bacterial genome content through distinct mechanisms over different timescales.', 'authors' => 'Croucher NJ, Coupland PG, Stevenson AE, Callendrello A, Bentley SD, Hanage WP', 'description' => 'Bacterial populations often consist of multiple co-circulating lineages. Determining how such population structures arise requires understanding what drives bacterial diversification. Using 616 systematically sampled genomes, we show that Streptococcus pneumoniae lineages are typically characterized by combinations of infrequently transferred stable genomic islands: those moving primarily through transformation, along with integrative and conjugative elements and phage-related chromosomal islands. The only lineage containing extensive unique sequence corresponds to a set of atypical unencapsulated isolates that may represent a distinct species. However, prophage content is highly variable even within lineages, suggesting frequent horizontal transmission that would necessitate rapidly diversifying anti-phage mechanisms to prevent these viruses sweeping through populations. Correspondingly, two loci encoding Type I restriction-modification systems able to change their specificity over short timescales through intragenomic recombination are ubiquitous across the collection. Hence short-term pneumococcal variation is characterized by movement of phage and intragenomic rearrangements, with the slower transfer of stable loci distinguishing lineages.', 'date' => '2014-11-19', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25407023', 'doi' => '', 'modified' => '2015-07-24 15:39:04', 'created' => '2015-07-24 15:39:04', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 8 => array( 'id' => '2838', 'name' => 'Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance', 'authors' => 'Mark R Davies, Matthew T Holden, Paul Coupland, Jonathan H K Chen, Carola Venturini, Timothy C Barnett, Nouri L Ben Zakour, Herman Tse, Gordon Dougan, Kwok-Yung Yuen & Mark J Walker', 'description' => '<p><span>A scarlet fever outbreak began in mainland China and Hong Kong in 2011 (refs. </span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref1" title="Chen, M. et al. Outbreak of scarlet fever associated with emm12 type group A Streptococcus in 2011 in Shanghai, China. Pediatr. Infect. Dis. J. 31, e158-e162 (2012)." id="ref-link-1">1</a><span>–</span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref6" title="Yang, P. et al. Characteristics of group A Streptococcus strains circulating during scarlet fever epidemic, Beijing, China, 2011. Emerg. Infect. Dis. 19, 909-915 (2013)." id="ref-link-2">6</a><span>). Macrolide- and tetracycline-resistant </span><i>Streptococcus pyogenes emm</i><span>12 isolates represent the majority of clinical cases. Recently, we identified two mobile genetic elements that were closely associated with </span><i>emm</i><span>12 outbreak isolates: the integrative and conjugative element ICE-</span><i>emm</i><span>12, encoding genes for tetracycline and macrolide resistance, and prophage </span><span class="mb">Φ</span><span>HKU.vir, encoding the superantigens SSA and SpeC, as well as the DNase Spd1 (ref. </span><a href="http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html#ref4" title="Tse, H. et al. Molecular characterization of the 2011 Hong Kong scarlet fever outbreak. J. Infect. Dis. 206, 341-351 (2012)." id="ref-link-3">4</a><span>). Here we sequenced the genomes of 141 </span><i>emm</i><span>12 isolates, including 132 isolated in Hong Kong between 2005 and 2011. We found that the introduction of several ICE-</span><i>emm</i><span>12 variants, </span><span class="mb">Φ</span><span>HKU.vir and a new prophage, </span><span class="mb">Φ</span><span>HKU.ssa, occurred in three distinct </span><i>emm</i><span>12 lineages late in the twentieth century. Acquisition of </span><i>ssa</i><span> and transposable elements encoding multidrug resistance genes triggered the expansion of scarlet fever–associated </span><i>emm</i><span>12 lineages in Hong Kong. The occurrence of multidrug-resistant </span><i>ssa</i><span>-harboring scarlet fever strains should prompt heightened surveillance within China and abroad for the dissemination of these mobile genetic elements.</span></p>', 'date' => '2014-11-17', 'pmid' => 'http://www.nature.com/ng/journal/v47/n1/full/ng.3147.html', 'doi' => '10.1038/ng.3147', 'modified' => '2016-03-08 14:33:49', 'created' => '2016-03-08 14:33:21', 'ProductsPublication' => array( [maximum depth reached] ) ), (int) 9 => array( 'id' => '2262', 'name' => 'Microevolution of Burkholderia pseudomallei during an acute infection.', 'authors' => 'Limmathurotsakul D, Holden MT, Coupland P, Price EP, Chantratita N, Wuthiekanun V, Amornchai P, Parkhill J, Peacock SJ', 'description' => 'We used whole-genome sequencing to evaluate 69 independent colonies of Burkholderia pseudomallei isolated from seven body sites of a patient with acute disseminated melioidosis. Fourteen closely related genotypes were found, providing evidence for the rapid in vivo diversification of B. pseudomallei after inoculation and systemic spread.', 'date' => '2014-09-01', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24966357', 'doi' => '', 'modified' => '2015-07-24 15:39:03', 'created' => '2015-07-24 15:39:03', 'ProductsPublication' => array( [maximum depth reached] ) ) ), 'Testimonial' => array( (int) 0 => array( 'id' => '57', 'name' => 'VIB - Megaruptor', 'description' => '<p>At the VIB Department of Molecular Genetics we strive to implement the best technologies in our workflows. Recent advances in long read sequencing using Oxford Nanopore technology have made the investigation of complex genomic variation in the human genome possible. A crucial step in the beginning of the library preparation is the shearing of genomic DNA, and this is where we implement the Megaruptor. The straightforward protocol is efficient, requires minimal sample handling and results in very reproducible fragment sizes. Another advantage is the consumable cost, which is lower than alternative technologies. For our applications we prefer shearing to lengths of 10 to 40kb, and for this the Megaruptor is the optimal solution.</p>', 'author' => 'Wouter de Coster and the Genomic Service Facility, VIB Department of Molecular Genetics, Antwerp, Belgium', 'featured' => true, 'slug' => 'vib-antwerp', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:35:57', 'created' => '2016-07-06 10:35:57', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 1 => array( 'id' => '42', 'name' => 'Norwegian Sequencing Center - Megaruptor', 'description' => '<div class="page" title="Page 1"> <div class="layoutArea"> <div class="column"> <p><span>The Norwegian sequencing centre (NSC) is a national core facility fully equipped to handle a diverse set of sequencing projects. We see an increased interest in long read technologies, in which our PacBio Single Molecule sequencing service is important. PacBio, (or SMRT) sequencing, like any other single molecule system, starts with a critical library preparation step to generate long fragment libraries of 10-20kb length. Even longer libraries can be made, but common to all library types is to start with good quality DNA of high molecular weight. Physical shearing of the DNA, targeting the desired library length, is the first critical step in the procedure. NSC has experience with different methods to fragment DNA like nebulization, enzymatic, tagmentation, covaris AFA, g-tubes (also covaris), custom syringe/needle protocols and hydroshearing. The best fragmentation method should produce a sharp fragmentation pattern close to the target length as defined by the protocol settings, wi</span><span>th as little “smear like” pattern of lower molecular length as possible. </span><span>The final step of PacBio library preparation is to size select only the longest fragments (> 7kb) as this increases the overall success rate of only sequencing the longest library fragments. It is not uncommon to loose a lot of material during this step. However, if the initial fragmentation yields sharp fragmentation patterns with minimal smear, more of the total DNA is retained in the final library. This is perhaps most important on samples were DNA amounts are limited. The optimal fragmentation method has to produce similar fragmentation pattern each time, with little deviation with sample type and input amount. We have found the new Megaruptor from Diagenode to be one of the best instruments for routine shearing of DNA to lengths of 2-40kb. It is easy to use, with walk away shearing, and the instrument does all the washes before, in between samples and after shearing without user intervention. Disposable hydropores is beneficial for lowering the contamination risk. The fact that it is so easy to use make it a good choice for laboratories with multiple users and/or when training new staff. </span></p> </div> </div> </div>', 'author' => 'Morten Skage and Dr Ave Tooming-Klunderud, Norwegian Sequencing Centre, Oslo, Norway', 'featured' => false, 'slug' => 'norwegian-sequencing-center-megaruptor', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-03-22 11:31:43', 'created' => '2015-11-26 09:21:28', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 2 => array( 'id' => '39', 'name' => 'Megaruptor® ', 'description' => '<p><span>We will use the Megaruptor<sup>®</sup> within our PacBio<sup>®</sup> Single Molecule Sequencing workflows, especially for the construction of very big insert (>20kb) libraries.</span></p>', 'author' => 'Andrea Patrignani, Anna Bratus and Ralph Schlapbach, Functional Genomics Center Zurich (ETHZ/UZH), Switzerland', 'featured' => false, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-11-23 10:04:40', 'created' => '2015-11-23 10:03:27', 'ProductsTestimonial' => array( [maximum depth reached] ) ), (int) 3 => array( 'id' => '3', 'name' => 'Megaruptor - Sylke Winkler', 'description' => '<p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p>', 'author' => 'Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany', 'featured' => true, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:38:08', 'created' => '2014-10-28 23:47:30', 'ProductsTestimonial' => array( [maximum depth reached] ) ) ), 'Area' => array(), 'SafetySheet' => array() ) $country = 'US' $countries_allowed = array( (int) 0 => 'CA', (int) 1 => 'US', (int) 2 => 'IE', (int) 3 => 'GB', (int) 4 => 'DK', (int) 5 => 'NO', (int) 6 => 'SE', (int) 7 => 'FI', (int) 8 => 'NL', (int) 9 => 'BE', (int) 10 => 'LU', (int) 11 => 'FR', (int) 12 => 'DE', (int) 13 => 'CH', (int) 14 => 'AT', (int) 15 => 'ES', (int) 16 => 'IT', (int) 17 => 'PT' ) $outsource = false $other_formats = array() $edit = '' $testimonials = '<blockquote><div class="page" title="Page 1"> <div class="layoutArea"> <div class="column"> <p><span>The Norwegian sequencing centre (NSC) is a national core facility fully equipped to handle a diverse set of sequencing projects. We see an increased interest in long read technologies, in which our PacBio Single Molecule sequencing service is important. PacBio, (or SMRT) sequencing, like any other single molecule system, starts with a critical library preparation step to generate long fragment libraries of 10-20kb length. Even longer libraries can be made, but common to all library types is to start with good quality DNA of high molecular weight. Physical shearing of the DNA, targeting the desired library length, is the first critical step in the procedure. NSC has experience with different methods to fragment DNA like nebulization, enzymatic, tagmentation, covaris AFA, g-tubes (also covaris), custom syringe/needle protocols and hydroshearing. The best fragmentation method should produce a sharp fragmentation pattern close to the target length as defined by the protocol settings, wi</span><span>th as little “smear like” pattern of lower molecular length as possible. </span><span>The final step of PacBio library preparation is to size select only the longest fragments (> 7kb) as this increases the overall success rate of only sequencing the longest library fragments. It is not uncommon to loose a lot of material during this step. However, if the initial fragmentation yields sharp fragmentation patterns with minimal smear, more of the total DNA is retained in the final library. This is perhaps most important on samples were DNA amounts are limited. The optimal fragmentation method has to produce similar fragmentation pattern each time, with little deviation with sample type and input amount. We have found the new Megaruptor from Diagenode to be one of the best instruments for routine shearing of DNA to lengths of 2-40kb. It is easy to use, with walk away shearing, and the instrument does all the washes before, in between samples and after shearing without user intervention. Disposable hydropores is beneficial for lowering the contamination risk. The fact that it is so easy to use make it a good choice for laboratories with multiple users and/or when training new staff. </span></p> </div> </div> </div><cite>Morten Skage and Dr Ave Tooming-Klunderud, Norwegian Sequencing Centre, Oslo, Norway</cite></blockquote> <blockquote><p><span>We will use the Megaruptor<sup>®</sup> within our PacBio<sup>®</sup> Single Molecule Sequencing workflows, especially for the construction of very big insert (>20kb) libraries.</span></p><cite>Andrea Patrignani, Anna Bratus and Ralph Schlapbach, Functional Genomics Center Zurich (ETHZ/UZH), Switzerland</cite></blockquote> ' $featured_testimonials = '<blockquote><span class="label-green" style="margin-bottom:16px;margin-left:-22px">TESTIMONIAL</span><p>At the VIB Department of Molecular Genetics we strive to implement the best technologies in our workflows. Recent advances in long read sequencing using Oxford Nanopore technology have made the investigation of complex genomic variation in the human genome possible. A crucial step in the beginning of the library preparation is the shearing of genomic DNA, and this is where we implement the Megaruptor. The straightforward protocol is efficient, requires minimal sample handling and results in very reproducible fragment sizes. Another advantage is the consumable cost, which is lower than alternative technologies. For our applications we prefer shearing to lengths of 10 to 40kb, and for this the Megaruptor is the optimal solution.</p><cite>Wouter de Coster and the Genomic Service Facility, VIB Department of Molecular Genetics, Antwerp, Belgium</cite></blockquote> <blockquote><span class="label-green" style="margin-bottom:16px;margin-left:-22px">TESTIMONIAL</span><p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p><cite>Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany</cite></blockquote> ' $testimonial = array( 'id' => '3', 'name' => 'Megaruptor - Sylke Winkler', 'description' => '<p>The Megaruptor® shows a <strong>more dense size</strong> distribution of sheared fragments, and a <strong>higher</strong> and <strong>more reproducible yield</strong>.</p>', 'author' => 'Sylke Winkler, Sylvia Clausing, Nicola Gscheidel, Yannick Duport, Eugene Myers and Andreas Dahl, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany, and Deep Sequencing Group, BIOTEC, TU Dresden, Germany', 'featured' => true, 'slug' => '', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2016-07-06 10:38:08', 'created' => '2014-10-28 23:47:30', 'ProductsTestimonial' => array( 'id' => '24', 'product_id' => '1824', 'testimonial_id' => '3' ) ) $related_products = '<li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydro-tubes-50-pc"><img src="/img/product/shearing_technologies/C30010018%20_hydro_tubes.png" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">C30010018</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2647" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2647" id="CartAdd/2647Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2647" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydro Tubes</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydro Tubes', 'C30010018', '65', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydro Tubes', 'C30010018', '65', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydro-tubes-50-pc" data-reveal-id="cartModal-2647" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydro Tubes</h6> </div> </div> </li> <li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydropore-short-10-pc"><img src="/img/product/shearing_technologies/hydropores.jpg" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">E07010001</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2645" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2645" id="CartAdd/2645Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2645" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydropore - short</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - short', 'E07010001', '160', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - short', 'E07010001', '160', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydropore-short-10-pc" data-reveal-id="cartModal-2645" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydropore - short</h6> </div> </div> </li> <li> <div class="row"> <div class="small-12 columns"> <a href="/en/p/hydropore-long-10-pc"><img src="/img/product/shearing_technologies/hydropores.jpg" alt="some alt" class="th"/></a> </div> <div class="small-12 columns"> <div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px"> <span class="success label" style="">E07010002</span> </div> <div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px"> <!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a--> <!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-2646" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog"> <form action="/en/carts/add/2646" id="CartAdd/2646Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="2646" id="CartProductId"/> <div class="row"> <div class="small-12 medium-12 large-12 columns"> <p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Hydropore - long</strong> to my shopping cart.</p> <div class="row"> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - long', 'E07010002', '170', $('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div> <div class="small-6 medium-6 large-6 columns"> <button class="alert small button expand" onclick="$(this).addToCart('Hydropore - long', 'E07010002', '170', $('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div> </div> </div> </div> </form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="hydropore-long-10-pc" data-reveal-id="cartModal-2646" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a> </div> </div> <div class="small-12 columns" > <h6 style="height:60px">Hydropore - long</h6> </div> </div> </li> ' $related = array( 'id' => '2646', 'antibody_id' => null, 'name' => 'Hydropore - long', 'description' => '<p><span style="font-weight: 400;">Hydropore-longは<strong>10 kb - 75 kb</strong>の再現性のあるDNA断片化を可能にします。HydroporesはMegaruptor®(B06010001)と併用する特別設計になっています。</span></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label1' => '特徴', 'info1' => '<p>Sterile hydropore are intended for single use</p> <p><strong>Storage</strong><br />Store at room temperature</p> <p><strong>Precautions</strong><br />This product is for research use only. Not for use in diagnostic or therapeutic procedures</p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label2' => '', 'info2' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'label3' => '', 'info3' => '<p></p> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script> <script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>', 'format' => '10 pc', 'catalog_number' => 'E07010002', 'old_catalog_number' => '', 'sf_code' => 'E07010002-', 'type' => 'ACC', 'search_order' => '01-Accessory', 'price_EUR' => '145', 'price_USD' => '170', 'price_GBP' => '130', 'price_JPY' => '22715', 'price_CNY' => '', 'price_AUD' => '425', 'country' => 'ALL', 'except_countries' => 'None', 'quote' => false, 'in_stock' => false, 'featured' => false, 'no_promo' => false, 'online' => true, 'master' => true, 'last_datasheet_update' => '0000-00-00', 'slug' => 'hydropore-long-10-pc', 'meta_title' => 'Hydropore - long', 'meta_keywords' => '', 'meta_description' => 'Hydropore - long', 'modified' => '2022-04-18 09:02:42', 'created' => '2015-06-29 14:08:20', 'ProductsRelated' => array( 'id' => '68', 'product_id' => '1824', 'related_id' => '2646' ), 'Image' => array( (int) 0 => array( 'id' => '113', 'name' => 'product/shearing_technologies/hydropores.jpg', 'alt' => 'some alt', 'modified' => '2015-06-10 17:28:55', 'created' => '2015-06-10 17:28:55', 'ProductsImage' => array( [maximum depth reached] ) ) ) ) $rrbs_service = array( (int) 0 => (int) 1894, (int) 1 => (int) 1895 ) $chipseq_service = array( (int) 0 => (int) 2683, (int) 1 => (int) 1835, (int) 2 => (int) 1836, (int) 3 => (int) 2684, (int) 4 => (int) 1838, (int) 5 => (int) 1839, (int) 6 => (int) 1856 ) $labelize = object(Closure) { } $old_catalog_number = '' $label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>' $document = array( 'id' => '783', 'name' => 'Megaruptor® DNA Shearing System User Manual v2', 'description' => '<div class="page" title="Page 4"> <div class="layoutArea"> <div class="column"> <p><span>The Megaruptor</span><span>® </span><span>was designed to provide researchers with a simple, automated, and reproducible device for the fragmentation of DNA in the range of 2 kb - 75 kb. Shearing performance is independent of the source, concentration, temperature, or salt content of a DNA sample. Our user-friendly software allows for two samples to be processed sequentially without additional user input and without cross-contamination. Just set the desired parameters and the automated system takes care of the rest. Clogging issues are eliminated in this design. </span></p> </div> </div> </div>', 'image_id' => null, 'type' => 'Manual', 'url' => 'files/products/shearing_technology/megaruptor/Megaruptor_manual.pdf', 'slug' => 'megaruptor-manual', 'meta_keywords' => '', 'meta_description' => '', 'modified' => '2015-07-23 17:29:32', 'created' => '2015-07-07 11:47:45', 'ProductsDocument' => array( 'id' => '656', 'product_id' => '1824', 'document_id' => '783' ) ) $publication = array( 'id' => '2262', 'name' => 'Microevolution of Burkholderia pseudomallei during an acute infection.', 'authors' => 'Limmathurotsakul D, Holden MT, Coupland P, Price EP, Chantratita N, Wuthiekanun V, Amornchai P, Parkhill J, Peacock SJ', 'description' => 'We used whole-genome sequencing to evaluate 69 independent colonies of Burkholderia pseudomallei isolated from seven body sites of a patient with acute disseminated melioidosis. Fourteen closely related genotypes were found, providing evidence for the rapid in vivo diversification of B. pseudomallei after inoculation and systemic spread.', 'date' => '2014-09-01', 'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24966357', 'doi' => '', 'modified' => '2015-07-24 15:39:03', 'created' => '2015-07-24 15:39:03', 'ProductsPublication' => array( 'id' => '203', 'product_id' => '1824', 'publication_id' => '2262' ) ) $externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/24966357" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755 View::_evaluate() - CORE/Cake/View/View.php, line 971 View::_render() - CORE/Cake/View/View.php, line 933 View::render() - CORE/Cake/View/View.php, line 473 Controller::render() - CORE/Cake/Controller/Controller.php, line 963 ProductsController::slug() - APP/Controller/ProductsController.php, line 1052 ReflectionMethod::invokeArgs() - [internal], line ?? Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491 Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193 Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167 [main] - APP/webroot/index.php, line 118
To ensure you see the information most relevant to you, please select your country. Please note that your browser will need to be configured to accept cookies.
Diagenode will process your personal data in strict accordance with its privacy policy. This will include sending you updates about us, our products, and resources we think would be of interest to you.

