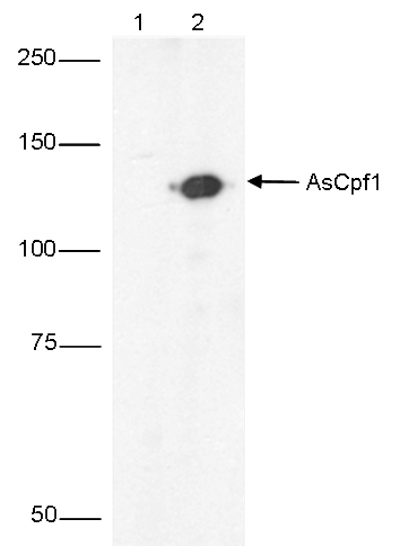
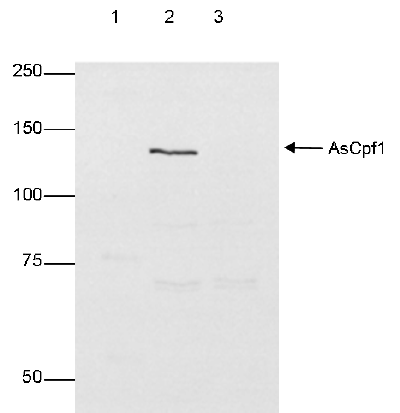
CRISPR systems are adaptable immune mechanisms which are present in many bacteria to protect themselves from foreign nucleic acids, such as viruses, transposable elements or plasmids. The CRISPR/Cas9 (CRISPR-associated protein 9nuclease) system from S. pyogenes was the first to be adapted for inducing sequence-specific double stranded breaks and targeted genome editing. This system is unique and flexible due to its dependence on RNA as the moiety that targets the nuclease to a desired DNA sequence and can be used to induce indel mutations, specific sequence replacements or insertions and large deletions or genomic rearrangements at any desired location in the genome. In addition, Cas9 can also be used to mediate upregulation of specific endogenous genes or to alter histone modifications or DNA methylation. Recently, a so-called type V CRISPR system has been identified in several bacteria which contains the Cpf1 (CRISPR from Prevotella and Francisella 1) protein. In contrast to Cas9 systems, CRISPR/Cpf1 systems do not require an additional trans-activating crRNA (tracrRNA), they cleave target DNA proceeded by a short T-rich protospacer-adjacent motif (PAM), in contrast to the G-rich PAM following the target DNA for Cas9, and they introduce a staggered DNA doublestranded break with a 4 or 5-nt 5’ overhang. Two of these CRISPR/Cpf1 systems, present in Acidaminococcus sp. and Lachnospiraceae bacterium have been identified as potential candidates for genome editing in mammalian cells.