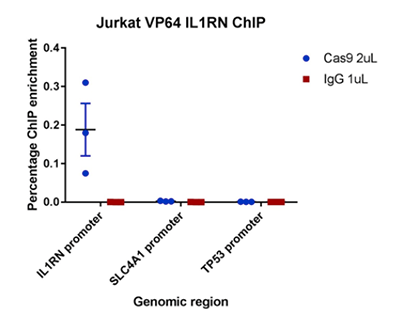
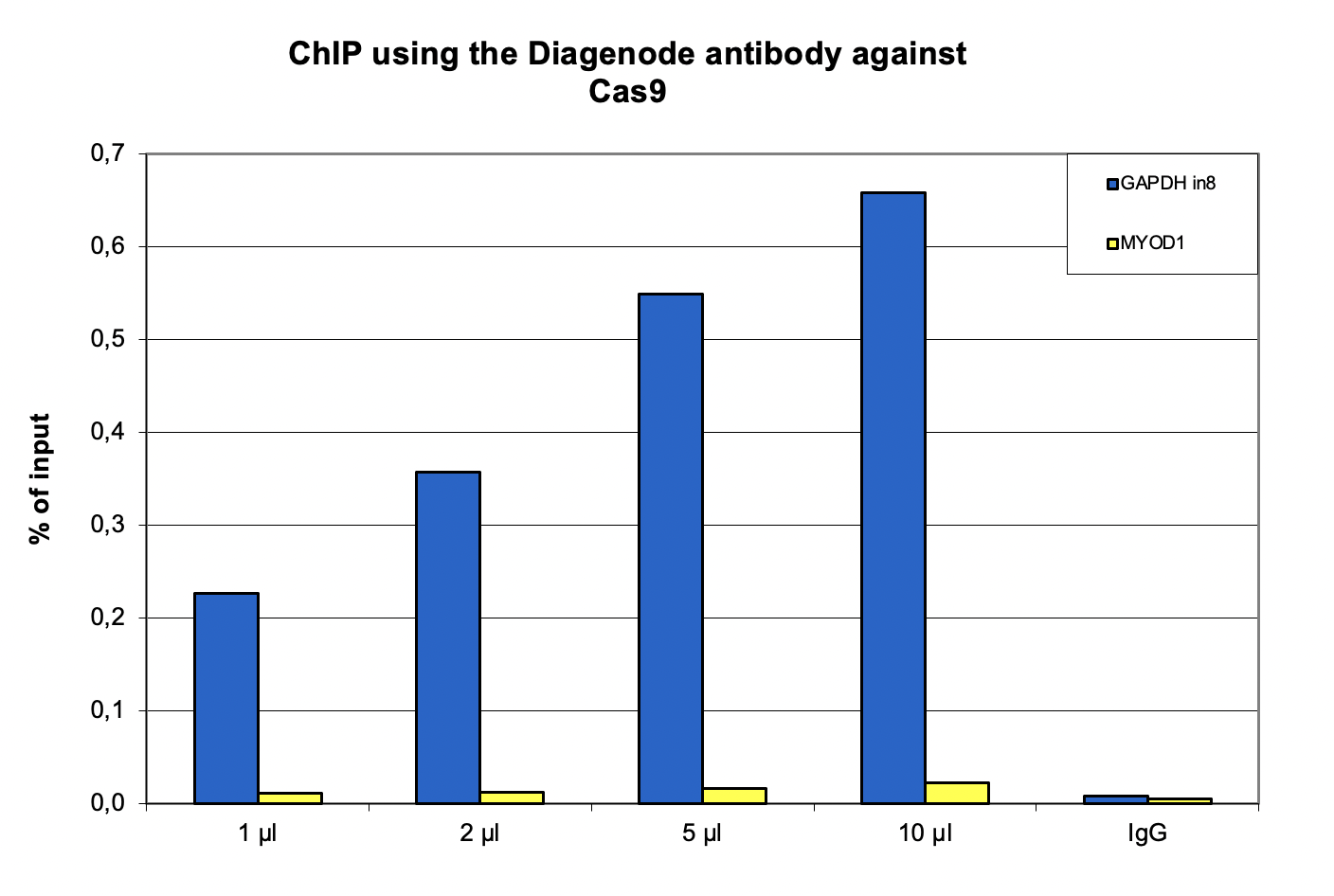
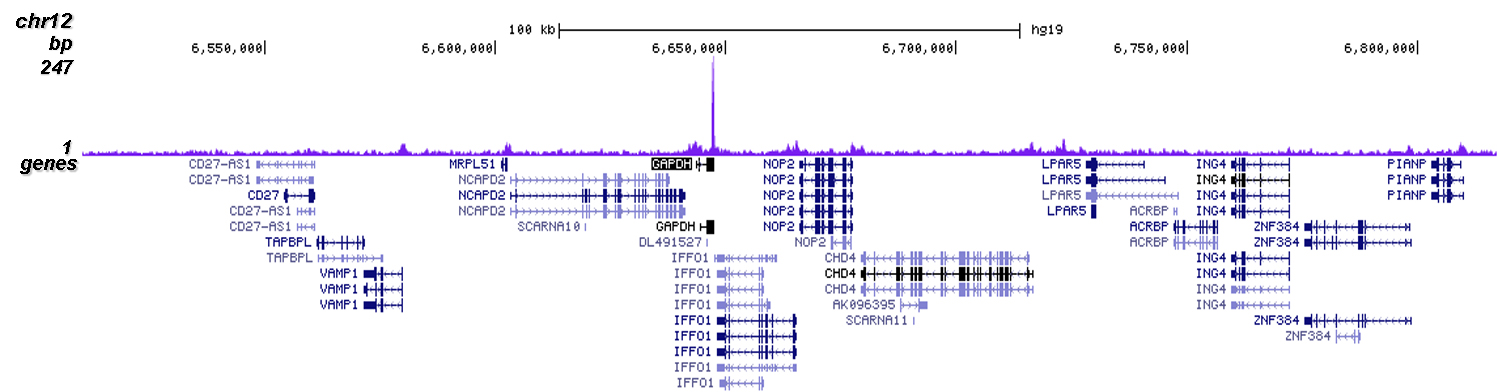
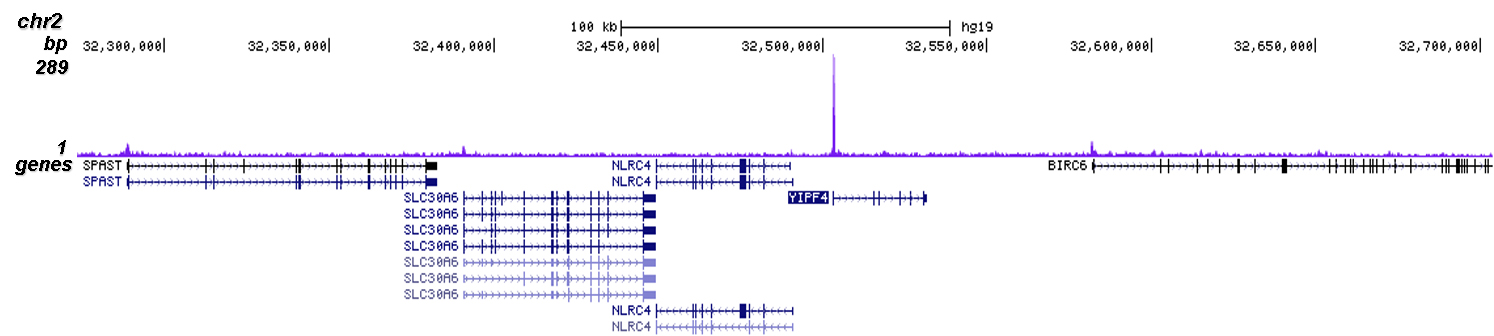
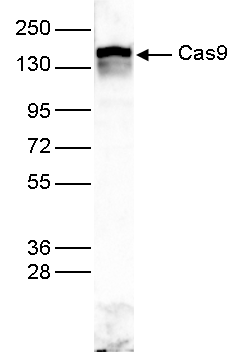
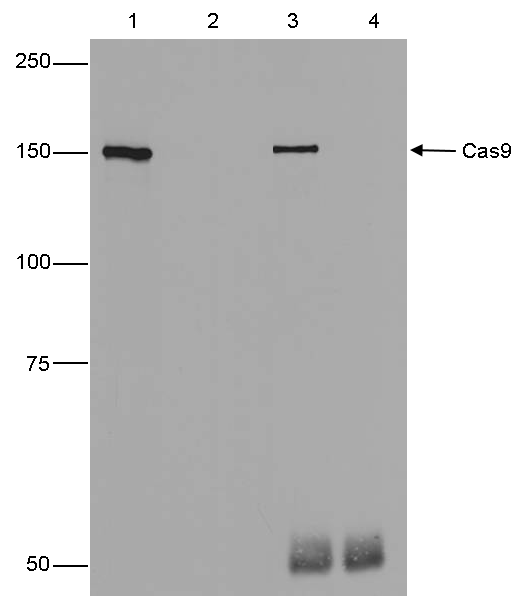
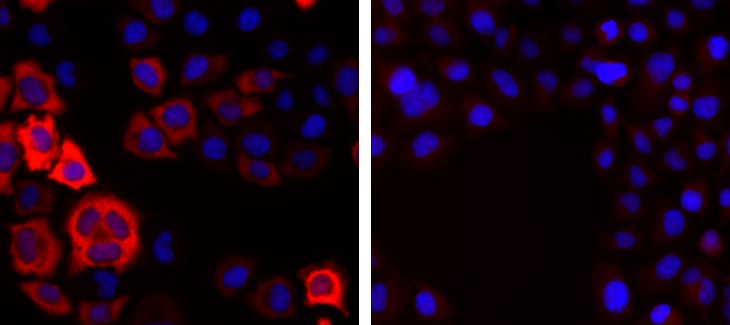
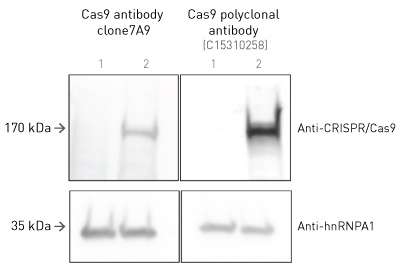
How to properly cite our product/service in your work We strongly recommend using this: CRISPR/Cas9 Antibody (Hologic Diagenode Cat# C15310258-100 Lot# A2508-004). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Transcriptional stress induces the overexpression of novel lncRNAs that regulate the BRCA1 locus
Samantha Cruz-Ruiz et al.
Long non-coding RNAs (lncRNAs) have been shown to play a role during transcriptional regulation in response to stress. However, their function under stress caused by transcriptional inhibition has not yet been addressed. Using genome-wide assays to elucidate the transcriptional response in human cells caused by RNA ... |
TurboCas: A method for locus-specific labeling of genomic regions and isolating their associated protein interactome
Bercin K Cenik et al.
Regulation of gene expression during development and stress response requires the concerted action of transcription factors and chromatin-binding proteins. Because this process is cell-type specific and varies with cellular conditions, mapping of chromatin factors at individual regulatory loci is crucial for underst... |
Human induced pluripotent stem cells for live cell cycle monitoring and endogenous gene activation
Kim R. et al.
The fluorescence ubiquitination cell cycle inhibitor (FUCCI) has been introduced to monitor cell cycle activity in living cells, including human induced pluripotent stem cells (hiPSC) and derived cell types. We have recently developed hiPSC with stable expression of dCas9VPR for endogenous gene activation and a Citr... |
Methyltransferase Inhibition Enables Tgf Driven Induction of and in Cancer Cells.
Liu Y-T et al.
deletion or silencing is common across human cancer, reinforcing the general importance of bypassing its tumor suppression in cancer formation or progression. In rhabdomyosarcoma (RMS) and neuroblastoma, two common childhood cancers, the three transcripts are independently expressed to varying degrees, but one, is u... |
Massively parallel multi-target CRISPR system interrogates Cas9-basedtarget recognition, DNA cleavage, and DNA repair
Zou Roger S. et al.
CRISPR-Cas9 nucleases, and particularly Streptococcus pyogenes Cas9, are widespread tools for genome editing. However, many aspects of intracellular Cas9 activity and the ensuing DNA damage response remain incompletely characterized. In order to address these issues, we developed a multiplexed CRISPR approach, where... |
Analysis of estrogen-regulated enhancer RNAs identifies a functionalmotif required for enhancer assembly and gene expression.
Hou Tim Y and Kraus W Lee
To better understand the functions of non-coding enhancer RNAs (eRNAs), we annotated the estrogen-regulated eRNA transcriptome in estrogen receptor α (ERα)-positive breast cancer cells using PRO-cap and RNA sequencing. We then cloned a subset of the eRNAs identified, fused them to single guide RNAs, and ... |
Avian influenza viruses suppress innate immunity by inducingtrans-transcriptional readthrough via SSU72.
Zhao Y. et al.
Innate immunity plays critical antiviral roles. The highly virulent avian influenza viruses (AIVs) H5N1, H7N9, and H5N6 can better escape host innate immune responses than the less virulent seasonal H1N1 virus. Here, we report a mechanism by which transcriptional readthrough (TRT)-mediated suppression of innate immu... |
Antisense non-coding transcription represses the <i>PHO5</i> model gene<i>via</i> remodelling of promoter chromatin structure
Novačić A. et al.
Pervasive transcription of eukaryotic genomes generates non-coding transcripts with regulatory potential. We examined the effects of non-coding antisense transcription on the regulation of expression of the yeast PHO5 gene, a paradigmatic case for gene regulation through promoter chromatin remodeling. By enhancing o... |
A predominant enhancer co-amplified with the oncogene is necessary andsufficient for its expression in squamous cancer
Liu Y. et al.
Amplification and overexpression of the SOX2 oncogene represent a hallmark of squamous cancers originating from diverse tissue types. Here, we find that squamous cancers selectively amplify a 3’ noncoding region together with SOX2, which harbors squamous cancer-specific chromatin accessible regions. We identif... |
Establishment of a second generation homozygous CRISPRa human inducedpluripotent stem cell (hiPSC) line for enhanced levels of endogenous geneactivation.
Schoger Eric et al.
CRISPR/Cas9 technology based on nuclease inactive dCas9 and fused to the heterotrimeric VPR transcriptional activator is a powerful tool to enhance endogenous transcription by targeting defined genomic loci. We generated homozygous human induced pluripotent stem cell (hiPSC) lines carrying dCas9 fused to VPR along w... |
Establishment of two homozygous CRISPR interference (CRISPRi)knock-in human induced pluripotent stem cell (hiPSC) lines for titratableendogenous gene repression.
Schoger Eric et al.
Using nuclease-deficient dead (d)Cas9 without enzymatic activity fused to transcriptional inhibitors (CRISPRi) allows for transcriptional interference and results in a powerful tool for the elucidation of developmental, homeostatic and disease mechanisms. We inserted dCas9KRAB (CRISPRi) cassette into the AAVS1 locus... |
TGFβ promotes widespread enhancer chromatin opening and operates ongenomic regulatory domains.
Guerrero-Martínez J. et al.
The Transforming Growth Factor-β (TGFβ) signaling pathway controls transcription by regulating enhancer activity. How TGFβ-regulated enhancers are selected and what chromatin changes are associated with TGFβ-dependent enhancers regulation are still unclear. Here we report that TGFβ treatment... |
Development of an ObLiGaRe Doxycycline Inducible Cas9 system for
pre-clinical cancer drug discovery.
Lundin, Anders and Porritt, Michelle J and Jaiswal, Himjyot and Seeliger,
Frank and Johansson, Camilla and Bidar, Abdel Wahad and Badertscher, Lukas
and Wimberger, Sandra and Davies, Emma J and Hardaker, Elizabeth and
Martins, Carla P and James, Emily and
The CRISPR-Cas9 system has increased the speed and precision of genetic
editing in cells and animals. However, model generation for drug
development is still expensive and time-consuming, demanding more target
flexibility and faster turnaround times with high reproducibility. The
generation of a tightly controlled O... |
A gene therapy for inherited blindness using dCas9-VPR–mediatedtranscriptional activation
Böhm, Sybille and Splith, Victoria and Riedmayr, Lisa Maria and Rötzer,René Dominik and Gasparoni, Gilles and Nordström, Karl J. V. and Wagner,Johanna Elisabeth and Hinrichsmeyer, Klara Sonnie and Walter, Jörn andWahl-Schott, Christian and Fenske, Stef
Catalytically inactive dCas9 fused to transcriptional activators (dCas9-VPR) enables activation of silent genes. Many disease genes have counterparts, which serve similar functions but are expressed in distinct cell types. One attractive option to compensate for the missing function of a defective gene could be to t... |
Guidelines for optimized gene knockout using CRISPR/Cas9
Campenhout CV et al.
CRISPR/Cas9 technology has evolved as the most powerful approach to generate genetic models both for fundamental and preclinical research. Despite its apparent simplicity, the outcome of a genome-editing experiment can be substantially impacted by technical parameters and biological considerations. Here, we present ... |
CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency.
Matharu N, Rattanasopha S, Tamura S, Maliskova L, Wang Y, Bernard A, Hardin A, Eckalbar WL, Vaisse C, Ahituv N
A wide range of human diseases result from haploinsufficiency, where the function of one of the two gene copies is lost. Here, we targeted the remaining functional copy of a haploinsufficient gene using CRISPR-mediated activation (CRISPRa) in and heterozygous mouse models to rescue their obesity phenotype. Transgeni... |
(Po)STAC (Polycistronic SunTAg modified CRISPR) enables live-cell and fixed-cell super-resolution imaging of multiple genes
Neguembor M.V. et al.
CRISPR/dCas9-based labeling has allowed direct visualization of genomic regions in living cells. However, poor labeling efficiency and signal-to-background ratio have limited its application to visualize genome organization using super-resolution microscopy. We developed (Po)STAC (Polycistronic SunTAg modified CRISP... |
A self-inactivating system for AAV-mediated in vivo base editing
Zuo Y. et al.
DNA base editors have been harnessed as an exciting therapeutic platform for human diseases and are rapidly progressing into human clinical trials. However, persistent expression of base editors delivered via adeno-associated virus (AAV) poses concerns with specificity and immunogenicity. Here we develop selfinactiv... |