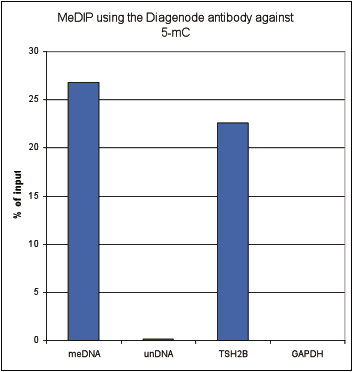
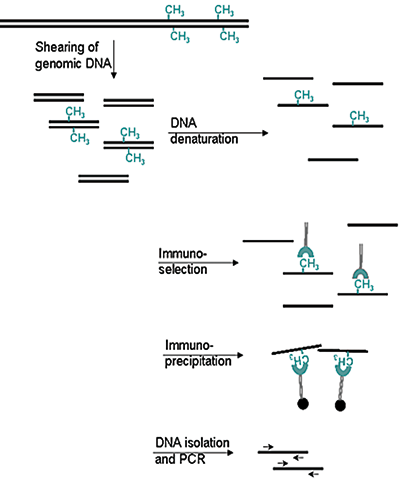
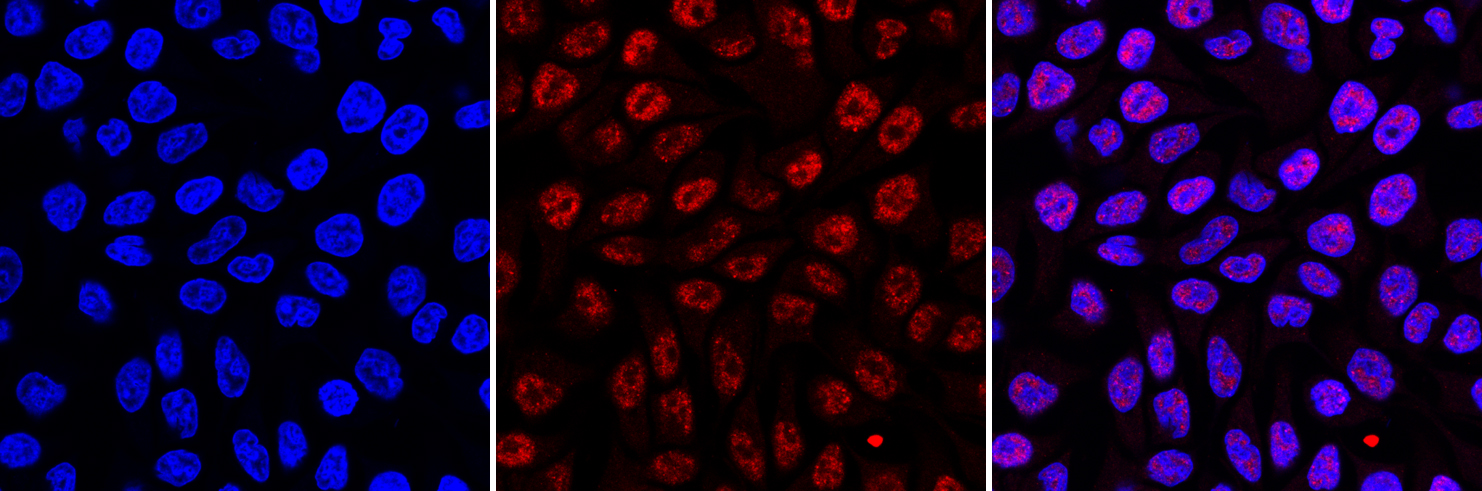
How to properly cite our product/service in your work We strongly recommend using this: 5-methylcytosine (5-mC) monoclonal antibody cl. b (sample size) (Hologic Diagenode Cat# C15200006-10 Lot# GF-005). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
An enriched maternal environment and stereotypies of sows differentiallyaffect the neuro-epigenome of brain regions related to emotionality intheir piglets.
Tatemoto P. et al.
Epigenetic mechanisms are important modulators of neurodevelopmental outcomes in the offspring of animals challenged during pregnancy. Pregnant sows living in a confined environment are challenged with stress and lack of stimulation which may result in the expression of stereotypies (repetitive behaviours without an... |
Role of epigenetics in the etiology of hypospadias through penileforeskin DNA methylation alterations.
Kaefer M. et al.
Abnormal penile foreskin development in hypospadias is the most frequent genital malformation in male children, which has increased dramatically in recent decades. A number of environmental factors have been shown to be associated with hypospadias development. The current study investigated the role of epigenetics i... |
Examination of Generational Impacts of Adolescent Chemotherapy:Ifosfamide and Potential for Epigenetic TransgenerationalInheritance
Thompson R. P. et al.
The current study was designed to use a rodent model to determine if exposure to the chemotherapy drug ifosfamide during puberty can induce altered phenotypes and disease in the grand-offspring of exposed individuals through epigenetic transgenerational inheritance. Pathologies such as delayed pubertal onset, kidney... |
Epigenome-wide association study of physical activity and physiologicalparameters in discordant monozygotic twins.
Duncan Glen E et al.
An epigenome-wide association study (EWAS) was performed on buccal cells from monozygotic-twins (MZ) reared together as children, but who live apart as adults. Cohorts of twin pairs were used to investigate associations between neighborhood walkability and objectively measured physical activity (PA) levels. Due to d... |
Environmental induced transgenerational inheritance impacts systemsepigenetics in disease etiology.
Beck D. et al.
Environmental toxicants have been shown to promote the epigenetic transgenerational inheritance of disease through exposure specific epigenetic alterations in the germline. The current study examines the actions of hydrocarbon jet fuel, dioxin, pesticides (permethrin and methoxychlor), plastics, and herbicides (glyp... |
GBS-MeDIP: A protocol for parallel identification of genetic andepigenetic variation in the same reduced fraction of genomes acrossindividuals.
Rezaei S. et al.
The GBS-MeDIP protocol combines two previously described techniques, Genotype-by-Sequencing (GBS) and Methylated-DNA-Immunoprecipitation (MeDIP). Our method allows for parallel and cost-efficient interrogation of genetic and methylomic variants in the DNA of many reduced genomes, taking advantage of the barcoding of... |
Preterm birth buccal cell epigenetic biomarkers to facilitatepreventative medicine.
Winchester P. et al.
Preterm birth is the major cause of newborn and infant mortality affecting nearly one in every ten live births. The current study was designed to develop an epigenetic biomarker for susceptibility of preterm birth using buccal cells from the mother, father, and child (triads). An epigenome-wide association study (EW... |
Epigenetic inheritance of DNA methylation changes in fish living inhydrogen sulfide-rich springs.
Kelley J. et al.
Environmental factors can promote phenotypic variation through alterations in the epigenome and facilitate adaptation of an organism to the environment. Although hydrogen sulfide is toxic to most organisms, the fish has adapted to survive in environments with high levels that exceed toxicity thresholds by orders of ... |
Epigenome-wide association study for pesticide (Permethrin and DEET)induced DNA methylation epimutation biomarkers for specifictransgenerational disease.
Thorson, Jennifer L M and Beck, Daniel and Ben Maamar, Millissia andNilsson, Eric E and Skinner, Michael K
BACKGROUND: Permethrin and N,N-diethyl-meta-toluamide (DEET) are the pesticides and insect repellent most commonly used by humans. These pesticides have been shown to promote the epigenetic transgenerational inheritance of disease in rats. The current study was designed as an epigenome-wide association study (EWAS) ... |
Between-Generation Phenotypic and Epigenetic Stability in a Clonal Snail.
Smithson, Mark and Thorson, Jennifer L M and Sadler-Riggleman, Ingrid andBeck, Daniel and Skinner, Michael K and Dybdahl, Mark
Epigenetic variation might play an important role in generating adaptive phenotypes by underpinning within-generation developmental plasticity, persistent parental effects of the environment (e.g., transgenerational plasticity), or heritable epigenetically based polymorphism. These adaptive mechanisms should be most... |
DNA methylation variation in the brain of laying hens in relation to differential behavioral patterns
Guerrero-Bosagna Carlos, Pértille Fábio, Gomez Yamenah, Rezaei Shiva, Gebhardt Sabine, Vögeli Sabine, Stratmann Ariane, Vöelkl Bernhard, Toscano Michael J.
Domesticated animals are unique to investigate the contribution of genetic and non-genetic factors to specific phenotypes. Among non-genetic factors involved in phenotype formation are epigenetic mechanisms. Here we aimed to identify whether relative DNA methylation differences in the nidopallium between groups of i... |
Sperm DNA Methylation Epimutation Biomarkers for Male Infertility and FSH Therapeutic Responsiveness.
Luján S, Caroppo E, Niederberger C, Arce JC, Sadler-Riggleman I, Beck D, Nilsson E, Skinner MK
Male factor infertility is increasing and recognized as playing a key role in reproductive health and disease. The current primary diagnostic approach is to assess sperm quality associated with reduced sperm number and motility, which has been historically of limited success in separating fertile from infertile male... |
Epigenetic transgenerational inheritance of parent-of-origin allelic transmission of outcross pathology and sperm epimutations
Ben Maamar Millissia, King Stephanie E., Nilsson Eric, Beck Daniel, Skinner Michael K.
Epigenetic transgenerational inheritance potentially impacts disease etiology, phenotypic variation, and evolution. An increasing number of environmental factors from nutrition to toxicants have been shown to promote the epigenetic transgenerational inheritance of disease. Previous observations have demonstrated tha... |
TET3 prevents terminal differentiation of adult NSCs by a non-catalytic action at Snrpn.
Montalbán-Loro R, Lozano-Ureña A, Ito M, Krueger C, Reik W, Ferguson-Smith AC, Ferrón SR
Ten-eleven-translocation (TET) proteins catalyze DNA hydroxylation, playing an important role in demethylation of DNA in mammals. Remarkably, although hydroxymethylation levels are high in the mouse brain, the potential role of TET proteins in adult neurogenesis is unknown. We show here that a non-catalytic action o... |
Environmental Toxicant Induced Epigenetic Transgenerational Inheritance of Prostate Pathology and Stromal-Epithelial Cell Epigenome and Transcriptome Alterations: Ancestral Origins of Prostate Disease.
Klukovich R, Nilsson E, Sadler-Riggleman I, Beck D, Xie Y, Yan W, Skinner MK
Prostate diseases include prostate cancer, which is the second most common male neoplasia, and benign prostatic hyperplasia (BPH), which affects approximately 50% of men. The incidence of prostate disease is increasing, and some of this increase may be attributable to ancestral exposure to environmental toxicants an... |
Genomic integrity of ground-state pluripotency.
Jafari N, Giehr P, Hesaraki M, Baas R, de Graaf P, Timmers HTM, Walter J, Baharvand H, Totonchi M
Pluripotent cells appear to be in a transient state during early development. These cells have the capability to transition into embryonic stem cells (ESCs). It has been reported that mouse pluripotent cells cultivated in chemically defined media sustain the ground state of pluripotency. Because the epigenetic patte... |
Developmental origins of transgenerational sperm DNA methylation epimutations following ancestral DDT exposure.
Ben Maamar M, Nilsson E, Sadler-Riggleman I, Beck D, McCarrey JR, Skinner MK
Epigenetic alterations in the germline can be triggered by a number of different environmental factors from diet to toxicants. These environmentally induced germline changes can promote the epigenetic transgenerational inheritance of disease and phenotypic variation. In previous studies, the pesticide DDT was shown ... |
Molecular Signatures of Regression of the Canine Transmissible Venereal Tumor.
Frampton D, Schwenzer H, Marino G, Butcher LM, Pollara G, Kriston-Vizi J, Venturini C, Austin R, de Castro KF, Ketteler R, Chain B, Goldstein RA, Weiss RA, Beck S, Fassati A
The canine transmissible venereal tumor (CTVT) is a clonally transmissible cancer that regresses spontaneously or after treatment with vincristine, but we know little about the regression mechanisms. We performed global transcriptional, methylation, and functional pathway analyses on serial biopsies of vincristine-t... |
Environmental toxicant induced epigenetic transgenerational inheritance of ovarian pathology and granulosa cell epigenome and transcriptome alterations: ancestral origins of polycystic ovarian syndrome and primary ovarian insufiency.
Nilsson E, Klukovich R, Sadler-Riggleman I, Beck D, Xie Y, Yan W, Skinner MK
Two of the most prevalent ovarian diseases affecting women's fertility and health are Primary Ovarian Insufficiency (POI) and Polycystic Ovarian Syndrome (PCOS). Previous studies have shown that exposure to a number of environmental toxicants can promote the epigenetic transgenerational inheritance of ovarian diseas... |