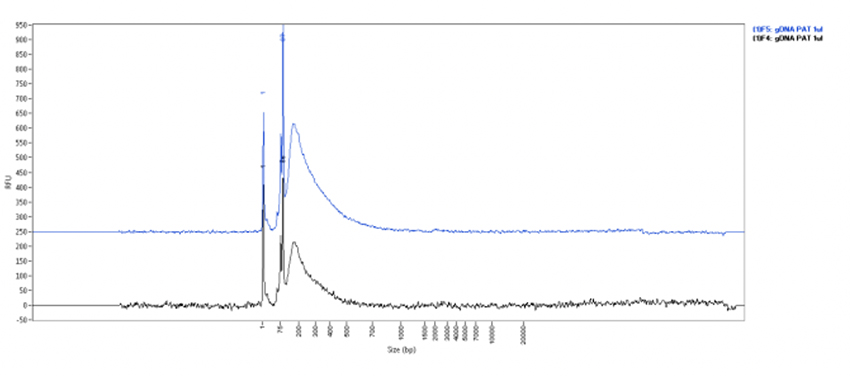
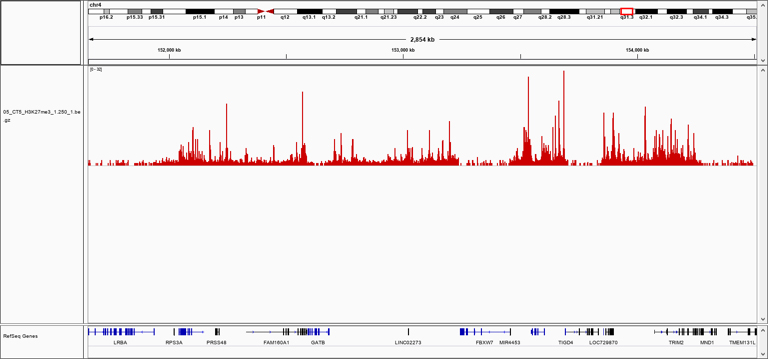
Figure 1: Quality control of pA-Tn5 transposase loaded with sequencing adapters
A: The Fragment Analyzer trace showing the representative cleavage pattern of gDNA. The pA-Tn5 fusion protein (Cat. No. C01070002) loaded with sequencing adapters efficiently digests gDNA to a smear. 500 ng of human genomic DNA were incubated for 7 min at 55°C with 1 μl of pATn5 fusion protein loaded with appropriated adaptors in a tagmentation buffer (40mM Tris-HCl pH7.5, 40mM MgCl2 and 12.5% DMF). The reaction was stopped by adding SDS, cleaned-up and resolved on the Fragment Analyzer to assess the cleavage.
B: Representative screenshot at selected locus obtained using Diagenode pA-Tn5 fusion protein (Cat. No. C01070002) lloaded with sequencing adapters and H3K27me3 polyclonal ChIP-seq grade antibody (Cat. No. C15410195) following CUT&Tag protocol (Kaya-Okur, H.S., Nat Commun 10, 1930 (2019)).
Protein A-Tn5 Is Compatible with Monoclonal Antibodies: High-Quality Profiling Demonstrated
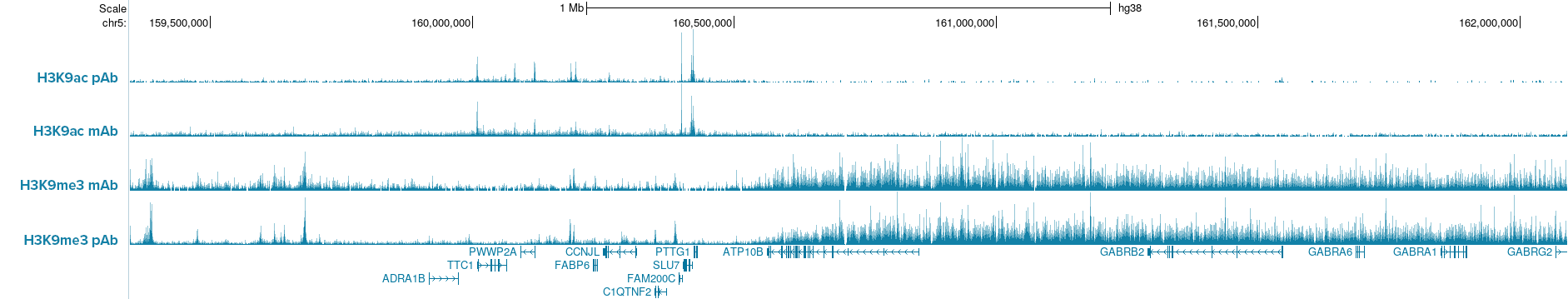
Figure 2. IGV browser tracks show CUT&Tag profiles of H3K9ac and H3K9me3 histone marks in K562 cells (50,000 cells per assay) using either monoclonal (mAb) or polyclonal (pAb) antibodies for each target. Comparable signal and peak enrichment demonstrate that Hologic Diagenode pA-Tn5 enables robust profiling with both monoclonal and polyclonal antibodies, indicating that pAG-Tn5 is not required. Antibodies used: H3K9ac pAb, Cat. no. C15410004; H3K9ac mAb, Cat. no. C152100185; H3K9me3 mAb, Cat. no. C15200146; H3K9me3 pAb, Cat. no. C15410056.


