CRISPR/Cas9
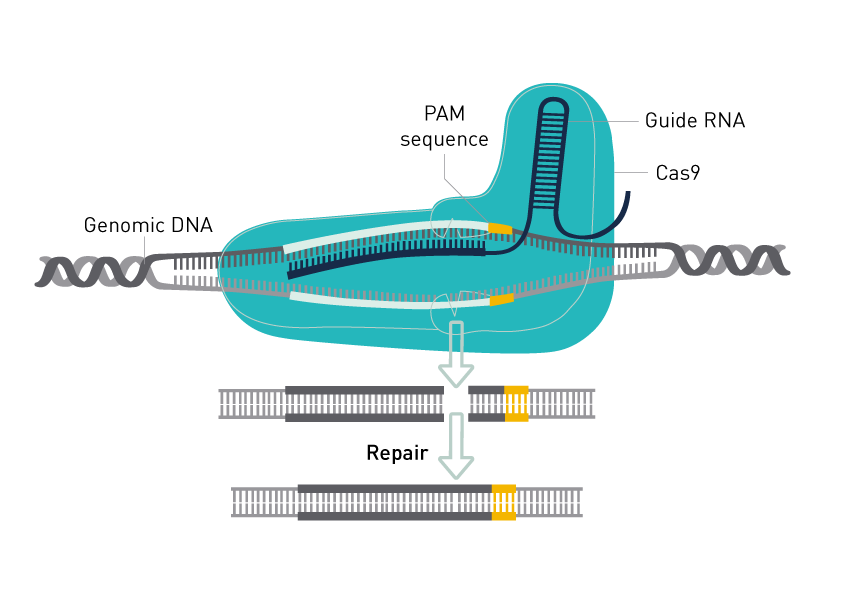
The CRISPR/Cas genome editing system uses a RNA-guided endonuclease to introduce site-directed double-strand breaks (DSBs) in DNA. The endonuclease Cas9 can be used for DNA cleavage at specific sites defined by guide RNAs and has been adopted as a technology of choice for genome engineering.
Cas9-mediated cleavage relies on the presence of a protospacer adjacent motif (PAM) in the target DNA. The Non Homologous End Joining (NHEJ) DNA repair pathway plays a role in repairing Cas9-induced DSBs. As the repair is prone to errors, insertions and deletions (indels) may occur that subsequently disrupt gene function. DSBs may be used to introduce specific sequence replacements such as insertions, large deletions, or genomic rearrangements at any genomic location.
Versions of Cas9 that bind but do not cleave DNA (dead Cas9) fused to specific catalytic domains can up- or down-regulate the expression of specific endogenous genes. In addition, they may alter histone modifications or DNA methylation.
Cas9 proteins from Streptococcus pyogenes and Staphylococcus aureus are the best characterized CRISPR-associated nucleases. Recently two new CRISPR-associated nucleases, Cpf1 from Lachnospiraceae bacterium and Cpf1 from Acidaminococcus sp., have been described.
New! Accurate QC to optimize CRISPR/Cas9 genome editing specificity
Check out our products

