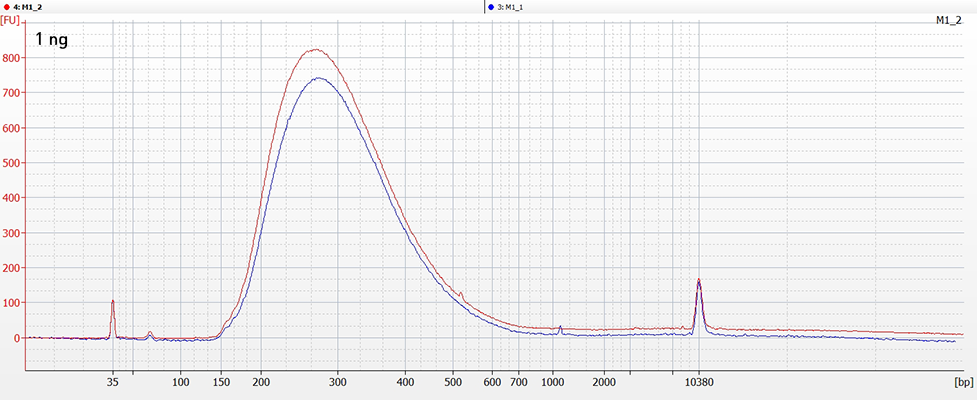
CATS relies on polynucleotide tailing for capturing the 3’-ends of nucleic acids and the template switching ability of MMLV-RT to capture the 5’ ends for cDNA synthesis. This kit allows for generating Illumina compatible ready-to-sequence libraries from RNA inputs as low as 100 picograms of polyA selected material in just few hours.
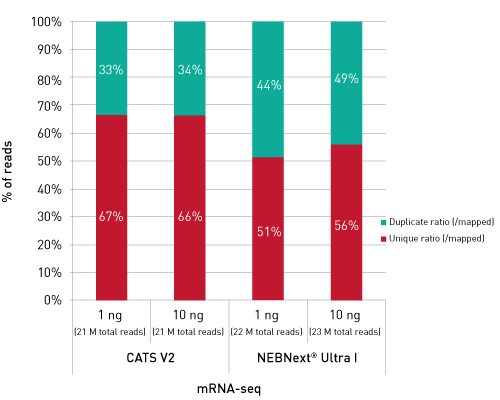
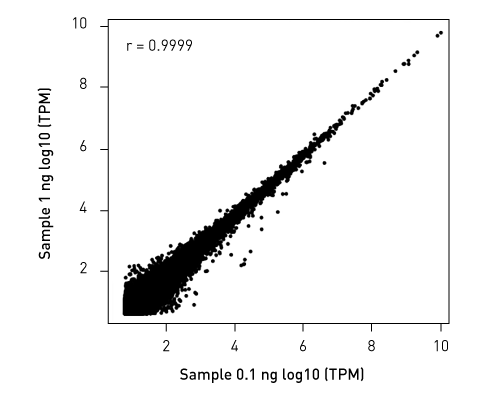
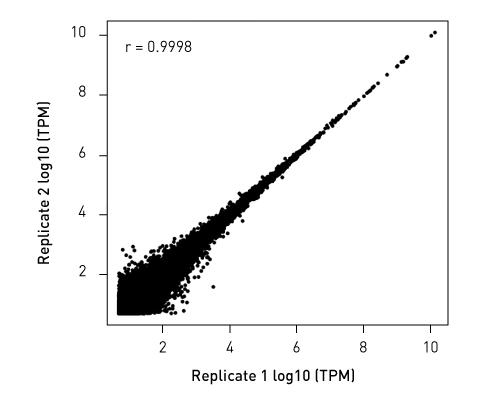
Libraries retain strand specificity of origin. Unlike competing solutions, our CATS RNA-seq kit exhibits higher library efficiency versus ligation-based methods providing higher complexity from better template capture and minimal bias due to lower amplification requirements.