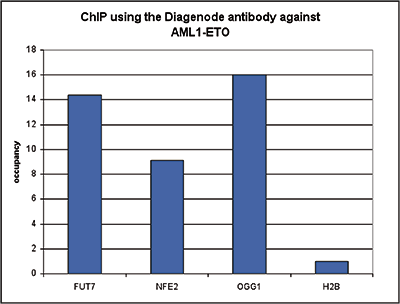
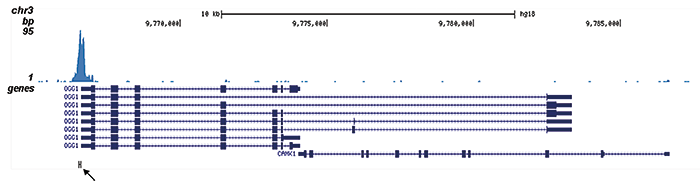
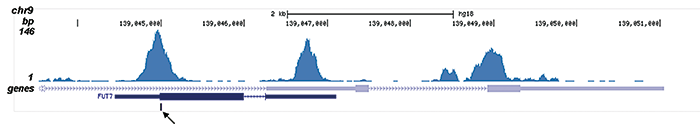
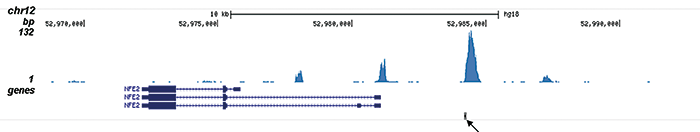
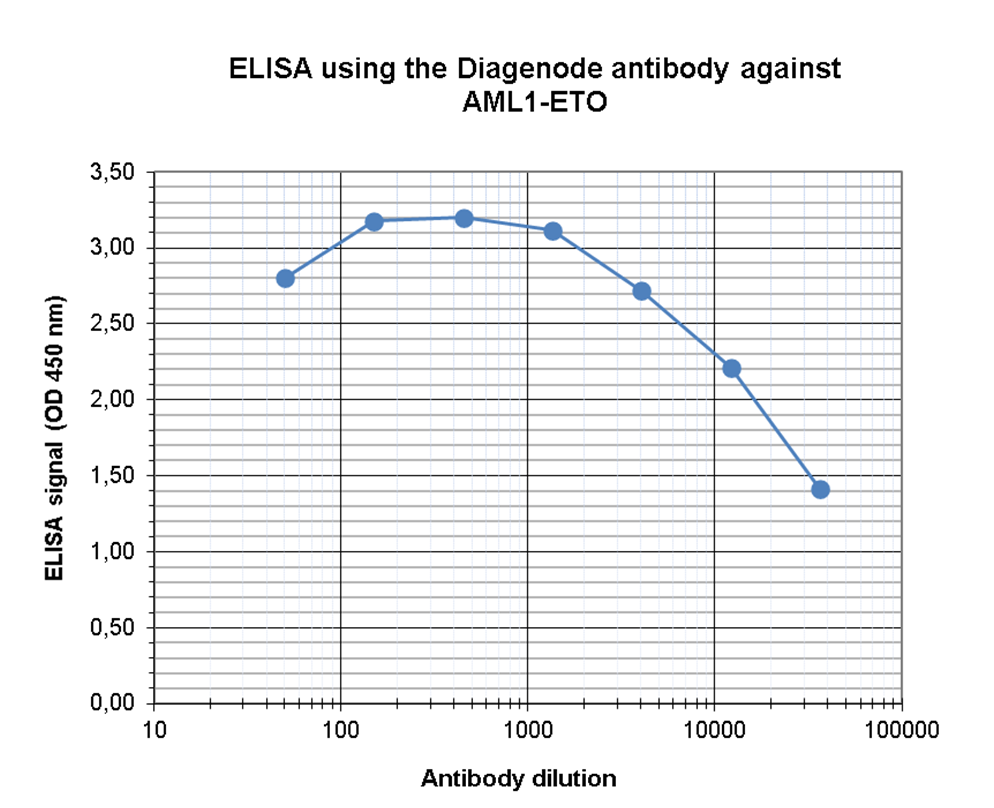
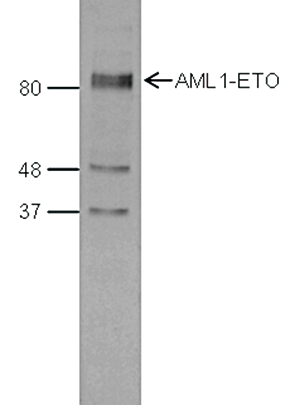
This antibody specifically recognizes the AML1 (RUNX1) (UniProtKB/Swiss-Prot entry Q01196) - ETO (RUNX1T1) (UniProtKB/ Swiss-Prot entry Q06455) fusion protein that arises due to a translocation between chromosome 8 and 22 (t(8;21)(q22;q22)). This translocation is one of the most frequent karyotypic abnormalities observed in acute myeloid leukaemia. It produces a chimerical gene made up of the 5’-region of AML1and the 3’-region of ETO. The chimerical protein is thought to associate with the nuclear corepressor/histone deacetylase complex to block hematopoietic differentiation.
-
产品
剪切技术
Tagmentation
Chromatin studies
DNA methylation
- Bisulfite conversion
- Methylated DNA Immunoprecipitation
- Methylbinding domain protein
- Hydroxymethylated DNA Immunoprecipitation
Genome editing (CRISPR/Cas9)
Antibodies
- All antibodies
- Sample size antibodies
- ChIP-seq grade antibodies
- ChIP-grade antibodies
- Western Blot Antibodies
- DNA modifications
- RNA modifications
- CRISPR/Cas9 antibodies
- CUT&Tag Antibodies
NGS Library preparation
- Library preparation for ChIP-seq
- Library preparation for RNA sequencing
- Library preparation for DNA sequencing
Automation
Reagents
- 服务
- 研究领域
- 资源
- 公司
-
联系人