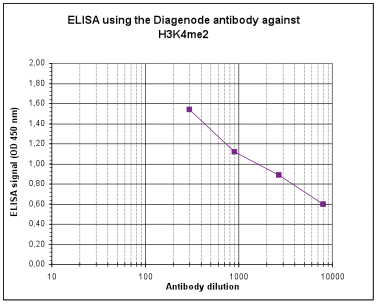
| ELISA Enzyme-linked immunosorbent assay. Read more |
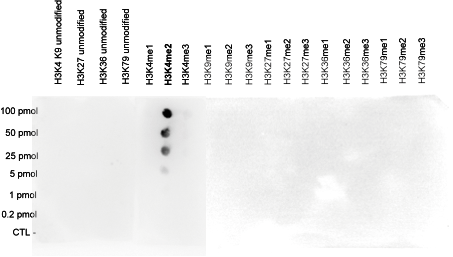
| DB Dot blotting Read more |
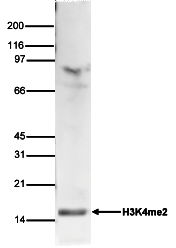
| WB Western blot : The quality of antibodies used in this technique is crucial for correct and specific protein identification. Diagenode offers huge selection of highly sensitive and specific western blot-validated antibodies. Learn more about: Load... Read more |
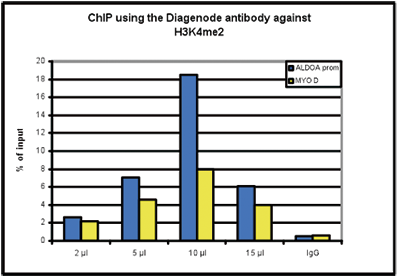
| ChIP-qPCR (ab) Read more |
-
产品
剪切技术
Tagmentation
Chromatin studies
DNA methylation
- Bisulfite conversion
- Methylated DNA Immunoprecipitation
- Methylbinding domain protein
- Hydroxymethylated DNA Immunoprecipitation
Genome editing (CRISPR/Cas9)
Antibodies
- All antibodies
- Sample size antibodies
- ChIP-seq grade antibodies
- ChIP-grade antibodies
- Western Blot Antibodies
- DNA modifications
- RNA modifications
- CRISPR/Cas9 antibodies
- CUT&Tag Antibodies
NGS Library preparation
- Library preparation for ChIP-seq
- Library preparation for RNA sequencing
- Library preparation for DNA sequencing
Automation
Reagents
- 服务
- 研究领域
- 资源
- 公司
-
联系人