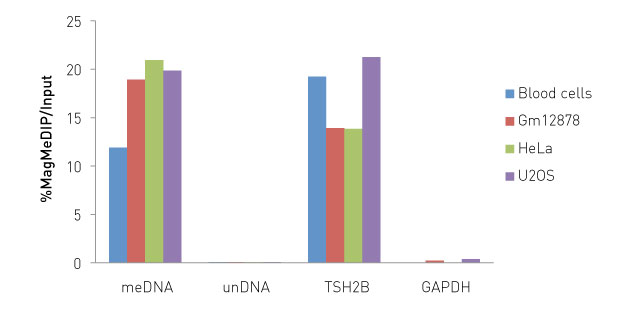
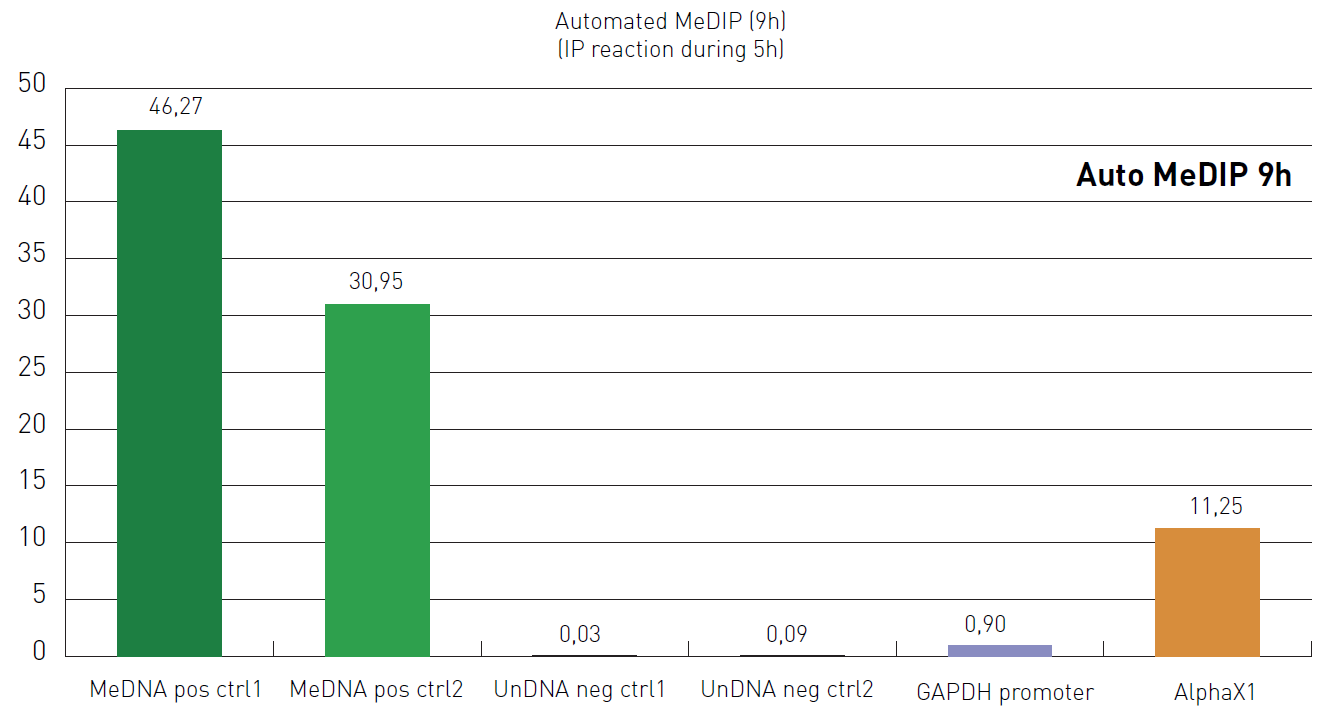
How to properly cite our product/service in your work We strongly recommend using this: Auto MagMeDIP qPCR Kit - ordering reference: C02010021 (Hologic Diagenode Cat# C02010014). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Human mitochondrial DNA is extensively methylated in a non-CpG context
Patil Vibha, Cuenin Cyrille, Chung Felicia, Aguilera Jesus R Rodriguez, Fernandez-Jimenez Nora, Romero-Garmendia Irati, Bilbao Jose Ramon, Cahais Vincent, Rothwell Joseph, Herceg Zdenko
Mitochondrial dysfunction plays critical roles in cancer development and related therapeutic response; however, exact molecular mechanisms remain unclear. Recently, alongside the discovery of mitochondrial-specific DNA methyltransferases, global and site-specific methylation of the mitochondrial genome has been desc... |
Protocols for Genetic and Epigenetic Studies of Rare Diseases Affecting Dental Tissues.
Amorim BR, Dos Santos PAC, de Lima CL, Andia DC, Mazzeu JF, Acevedo AC
This chapter describes methods related to the diagnosis of genetic dental diseases. Based on the present knowledge, clinical phenotyping and next-generation sequencing techniques are discussed. Methods necessary for Sanger sequencing, multiplex ligation-dependent probe amplification, and epigenetic modification meth... |
Molecular Signatures of Regression of the Canine Transmissible Venereal Tumor.
Frampton D, Schwenzer H, Marino G, Butcher LM, Pollara G, Kriston-Vizi J, Venturini C, Austin R, de Castro KF, Ketteler R, Chain B, Goldstein RA, Weiss RA, Beck S, Fassati A
The canine transmissible venereal tumor (CTVT) is a clonally transmissible cancer that regresses spontaneously or after treatment with vincristine, but we know little about the regression mechanisms. We performed global transcriptional, methylation, and functional pathway analyses on serial biopsies of vincristine-t... |
Saliva as a Blood Alternative for Genome-Wide DNA Methylation Profiling by Methylated DNA Immunoprecipitation (MeDIP) Sequencing
Staunstrup N.H. et al.
Background: Interrogation of DNA methylation profiles hold promise for improved diagnostics, as well as the delineation of the aetiology for common human diseases. However, as the primary tissue of the disease is often inaccessible without complicated and inconvenient interventions, there is an increasing interest i... |
Evaluating the Feasibility of DNA Methylation Analyses Using Long-Term Archived Brain Formalin-Fixed Paraffin-Embedded Samples
Bak S.T. et al.
We here characterize the usability of archival formalin-fixed paraffin-embedded (FFPE) brain tissue as a resource for genetic and DNA methylation analyses with potential relevance for brain-manifested diseases. We analyzed FFPE samples from The Brain Collection, Aarhus University Hospital Risskov, Denmark (AUBC), co... |
Genome-wide DNA methylation profiling with MeDIP-seq using archived dried blood spots
Nicklas H. Staunstrup et al.
Background In utero and early-life experienced environmental exposures are suggested to play an important role in many multifactorial diseases potentially mediated through lasting effects on the epigenome. As the epigenome in addition remains modifiable throughout life, identifying specific disease-relevant biomarke... |
Genome-wide non-CpG methylation of the host genome during M. tuberculosis infection
Sharma G et al.
A mammalian cell utilizes DNA methylation to modulate gene expression in response to environmental changes during development and differentiation. Aberrant DNA methylation changes as a correlate to diseased states like cancer, neurodegenerative conditions and cardiovascular diseases have been documented. Here we sho... |