- Fast & sensitive capture of methylated DNA
- High capture efficiency
- Differential fractionation of methylated DNA by CpG density (3 eluted fractions)
- Automation compatibility
MBD-seq allows for detection of genomic regions with different CpG density
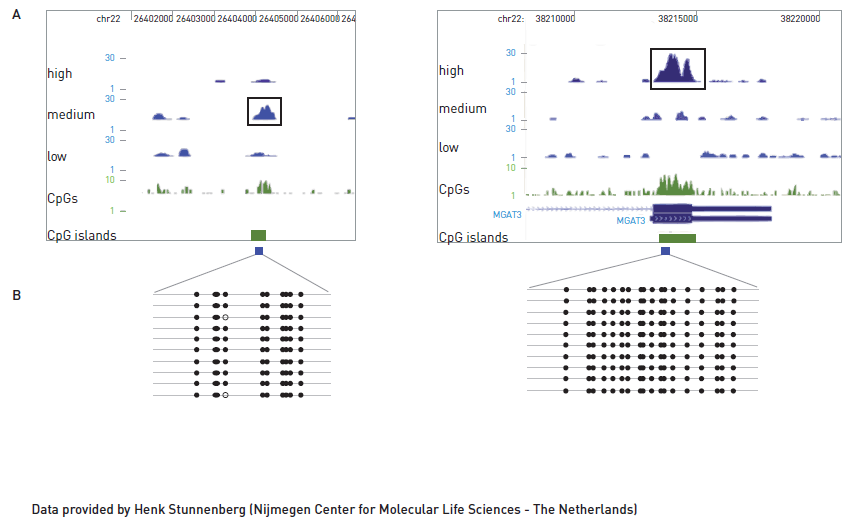
Figure 1. Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).

