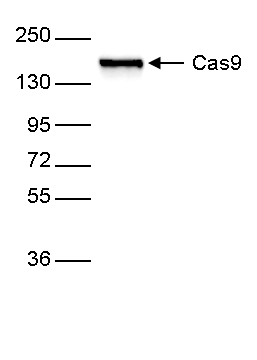
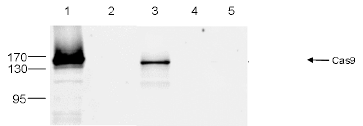
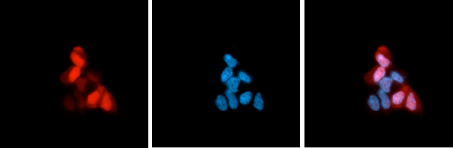
How to properly cite our product/service in your work We strongly recommend using this: CRISPR/Cas9 Antibody 4G10 (Hologic Diagenode Cat# C15200216-50 Lot# 004). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Dissecting the epigenetic regulation of the fetal hemoglobin genes to unravel a novel therapeutic approach for β-hemoglobinopathies
Amistadi, Simone et al.
Beta-hemoglobinopathies are severe genetic diseases caused by mutations affecting the production of the adult β-globin chain. The clinical severity is mitigated by the co-inheritance of mutations that reactivate the production of the fetal β-like γ-globin in adults. However, the epigenetic mechan... |
Cis-regulatory interfaces reveal the molecular mechanisms underlying the notochord gene regulatory network of Ciona
Negrón-Piñeiro L. J. et al.
Tissue-specific gene expression is fundamental in development and evolution, and is mediated by transcription factors and by the cis-regulatory regions (enhancers) that they control. Transcription factors and their respective tissue-specific enhancers are essential components of gene regulatory networks respons... |
Monitoring autochthonous lung tumors induced by somatic CRISPR geneediting in mice using a secreted luciferase.
Merle N. et al.
BACKGROUND: In vivo gene editing of somatic cells with CRISPR nucleases has facilitated the generation of autochthonous mouse tumors, which are initiated by genetic alterations relevant to the human disease and progress along a natural timeline as in patients. However, the long and variable, orthotopic tumor growth ... |
Systematic comparison of CRISPR-based transcriptional activatorsuncovers gene-regulatory features of enhancer-promoter interactions.
Wang K. et al.
Nuclease-inactivated CRISPR/Cas-based (dCas-based) systems have emerged as powerful technologies to synthetically reshape the human epigenome and gene expression. Despite the increasing adoption of these platforms, their relative potencies and mechanistic differences are incompletely characterized, particularly at h... |
DNA Methylation Editing by CRISPR-guided Excision of 5-Methylcytosine.
Devesa-Guerra I, Morales-Ruiz T, Pérez-Roldán J, Parrilla-Doblas JT, Dorado-León M, García-Ortiz MV, Ariza RR, Roldán-Arjona T
Tools for actively targeted DNA demethylation are required to increase our knowledge about regulation and specific functions of this important epigenetic modification. DNA demethylation in mammals involves TET-mediated oxidation of 5-methylcytosine (5-meC), which may promote its replication-dependent dilution and/or... |
Guidelines for optimized gene knockout using CRISPR/Cas9
Campenhout CV et al.
CRISPR/Cas9 technology has evolved as the most powerful approach to generate genetic models both for fundamental and preclinical research. Despite its apparent simplicity, the outcome of a genome-editing experiment can be substantially impacted by technical parameters and biological considerations. Here, we present ... |
Optimization of CRISPR/Cas9 Delivery to Human Hematopoietic Stem and Progenitor Cells for Therapeutic Genomic Rearrangements.
Lattanzi A, Meneghini V, Pavani G, Amor F, Ramadier S, Felix T, Antoniani C, Masson C, Alibeu O, Lee C, Porteus MH, Bao G, Amendola M, Mavilio F, Miccio A
Editing the β-globin locus in hematopoietic stem cells is an alternative therapeutic approach for gene therapy of β-thalassemia and sickle cell disease. Using the CRISPR/Cas9 system, we genetically modified human hematopoietic stem and progenitor cells (HSPCs) to mimic the large rearrangements in the &beta... |
Optimization of CRISPR/Cas9 genome editing for loss-of-function in the early chick embryo
Gandhi S. et al.
The advent of CRISPR/Cas9 has made genome editing possible in virtually any organism, including those not previously amenable to genetic manipulations. Here, we present an optimization of CRISPR/Cas9 for application to early avian embryos with improved efficiency via a three-fold strategy. First, we employed Cas9 pr... |
Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA
Liang W. et al.
While CRISPR-based gene knock out in mammalian cells has proven to be very efficient, precise insertion of genetic elements via the cellular homology directed repair (HDR) pathway remains a rate-limiting step to seamless genome editing. Under the conditions described here, we achieved up to 56% targeted integration ... |
CRISPR-Mediated Gene Targeting of Human Induced Pluripotent Stem Cells
Susan M. Byrne, George M. Church
CRISPR/Cas9 nuclease systems can create double-stranded DNA breaks at specific sequences to efficiently and precisely disrupt, excise, mutate, insert, or replace genes. However, human embryonic stem cells and induced pluripotent stem cells (iPSCs) are more difficult to transfect and less resilient to DNA damage than... |