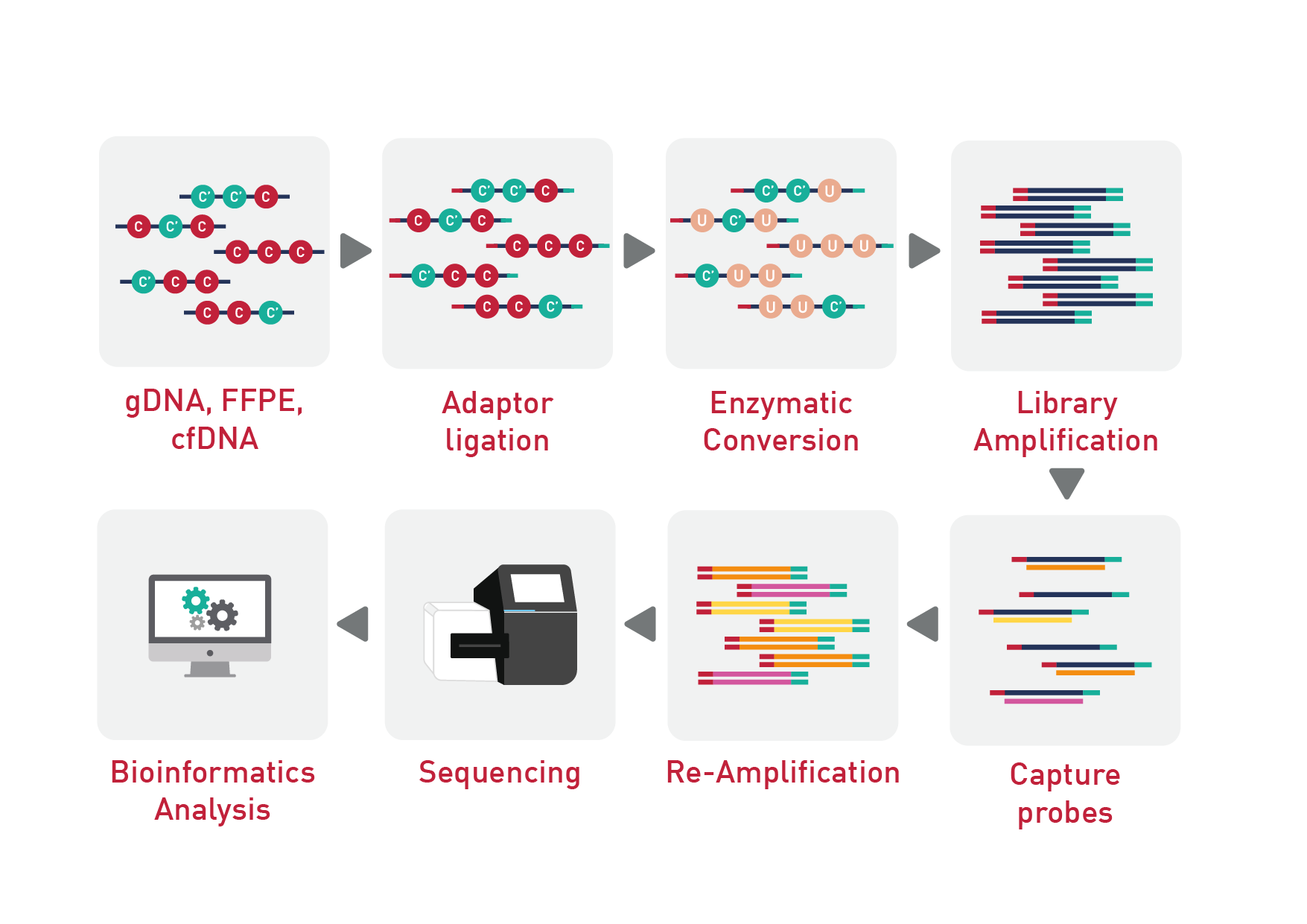
The Human Methylome Panel enables the identification and study of methylation biomarkers spanning a wide range of targets and applications. The 123 Mb panel provides a targeted NGS-based DNA methylation assay, covering more than 84% of human CpG island sites, which is 4 times the number of CpGs than other technologies and much more cost effective than WGBS. The high capture efficiency increases the sensitivity of detection and achieves a depth of coverage of 90% of bases at 30x coverage with high probe specificities.
- Optimal service for biomarker discovery in cancer, neurodegenerative, cardiovascular and metabolic diseases
- Ideal for challenging clinical samples such as liquid biopsies (plasma cfDNA) or FFPE
Workflow