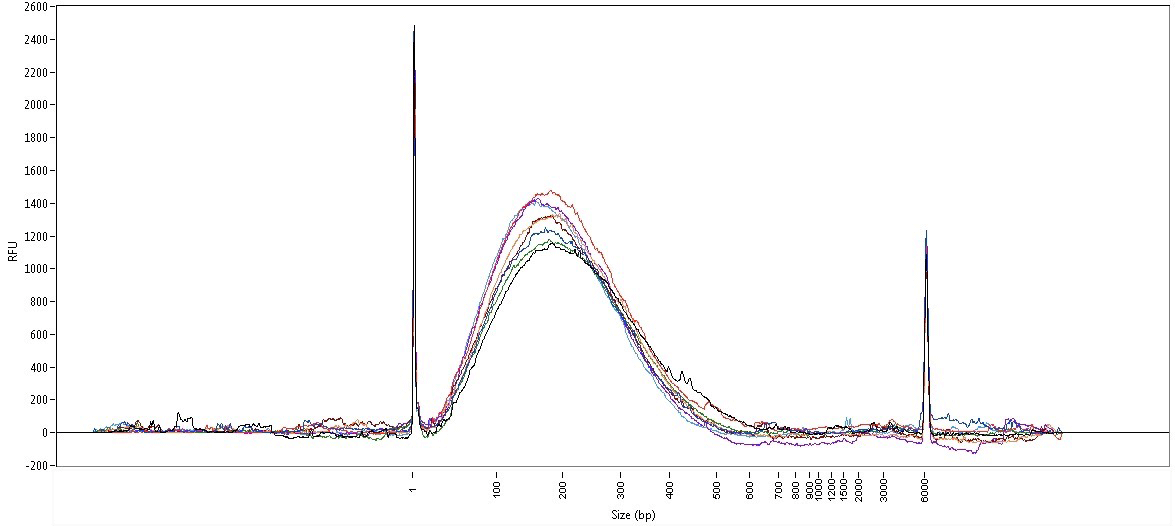
The One is specifically designed to prepare high-quality sheared DNA prior to create unbiaised libraries with even distribution and representation of the genome. The perfect unbiased shearing enables ultra-high and uniform coverage of libraries prepared from even AT-GC (E. Coli), AT-rich (S. Epidermis) and, GC-rich (B. Pertussis) bacterial genomes. The whole-genome sequencing on the Illumina® platform (Hiseq instrument, 50 base long single end reads) shows an excellent distribution for all three genomes.
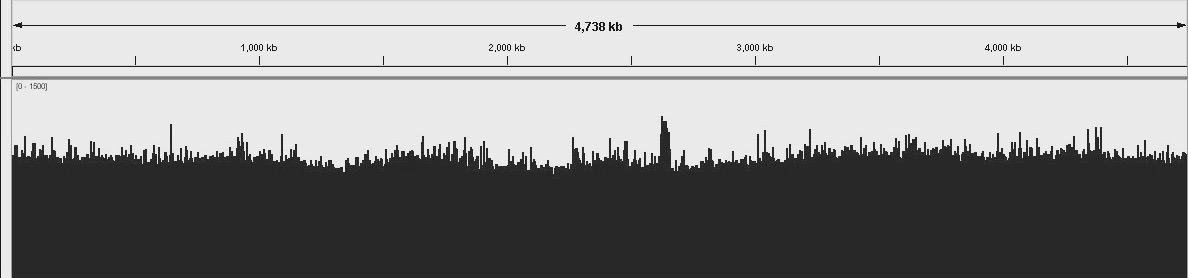
E. Coli genome uniform and consitent coverage. Number of matched reads to the reference genome.
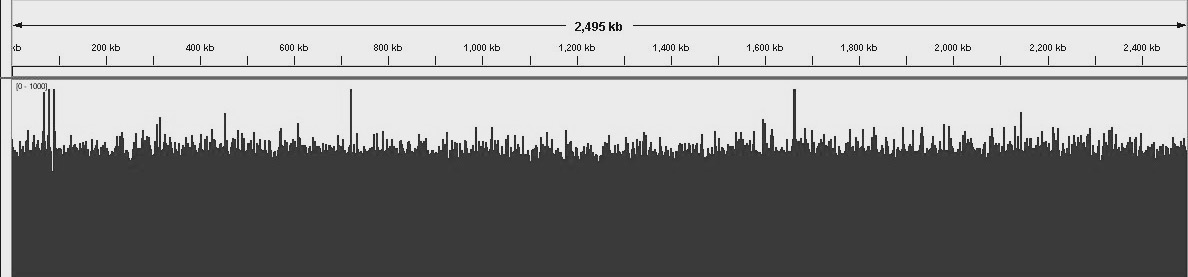
S. Epidermis genome uniform and consitent coverage. Number of matched reads to the reference genome.
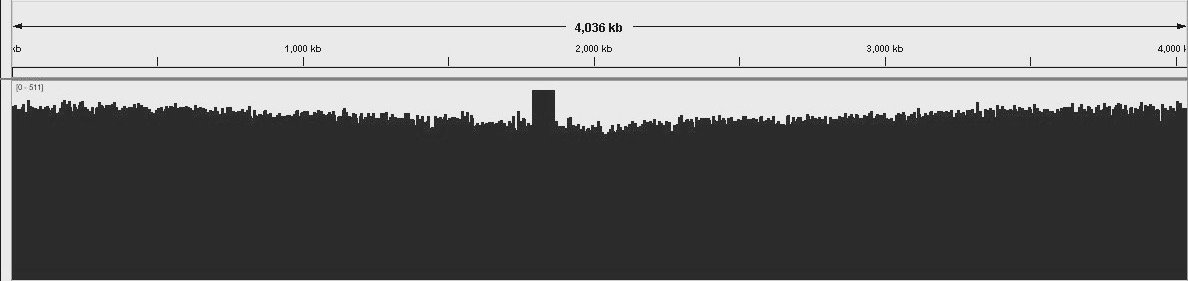
B. Pertussis genome uniform and consitent coverage. Number of matched reads to the reference genome.
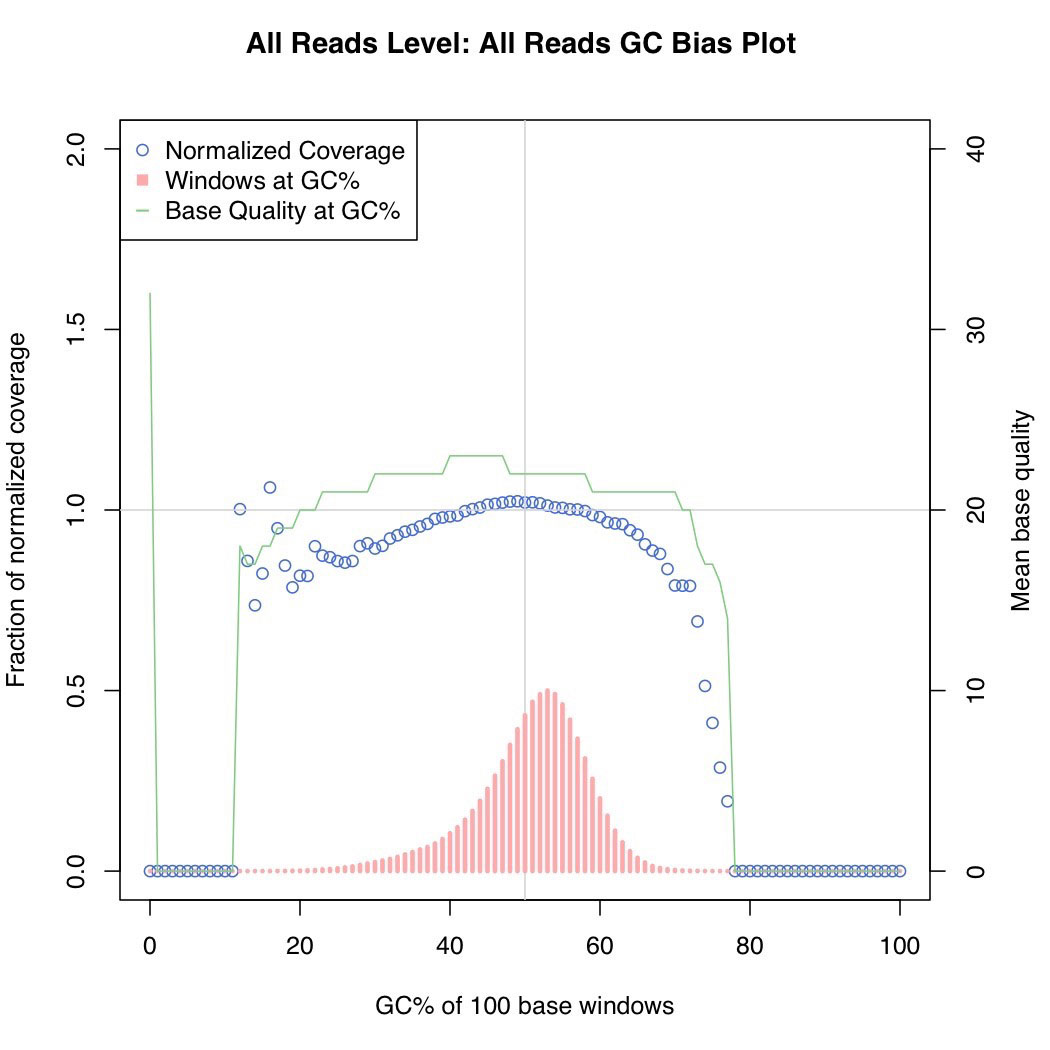
Unbiaised E. Coli DNA shearing for best coverage. Overall distribution of read depths for an even AT-GC genome.
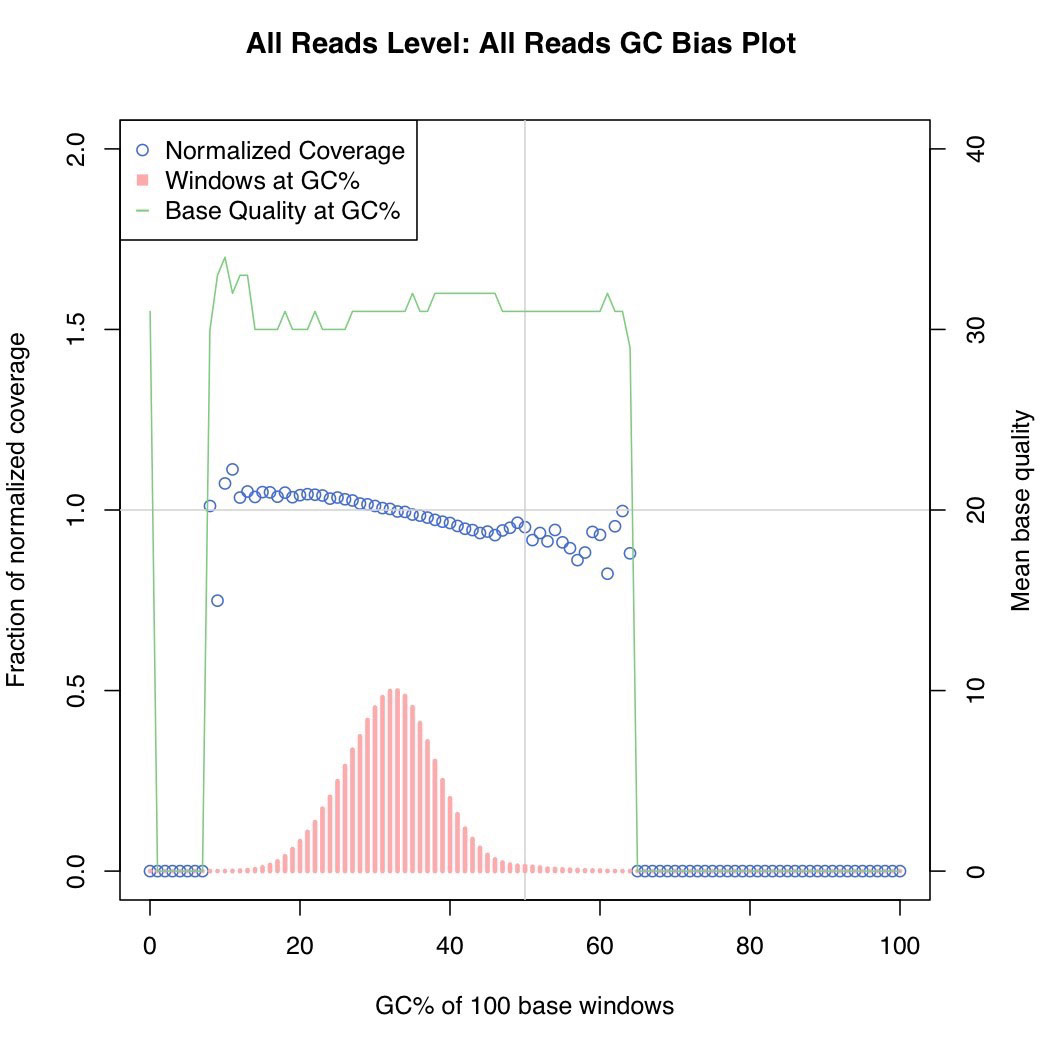
Unbiaised S. Epidermis DNA shearing for best coverage. Overall distribution of read depths for a AT-rich genome.
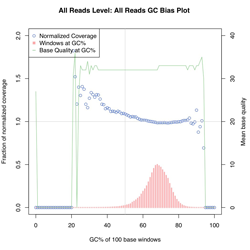
Unbiaised B. Pertussis DNA shearing for best coverage. Overall distribution of read depths for a CG-rich genome.