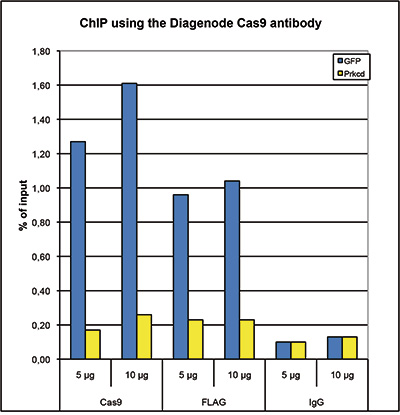
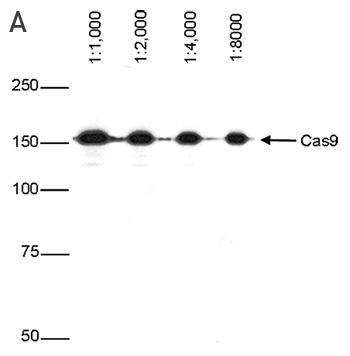
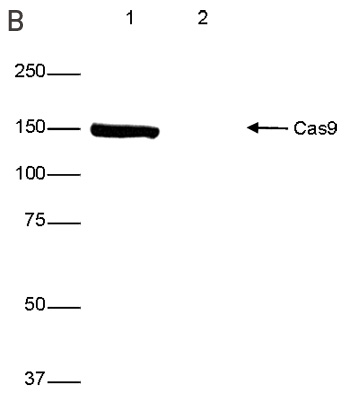
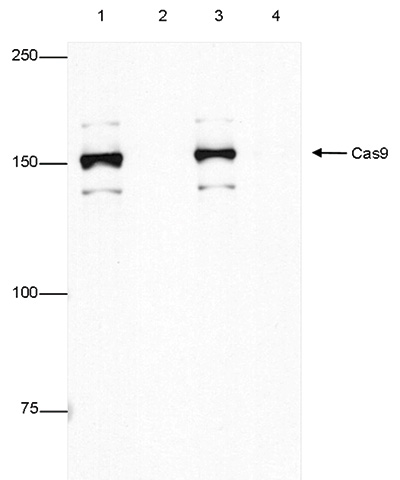
How to properly cite our product/service in your work We strongly recommend using this: CRISPR/Cas9 Antibody (Hologic Diagenode Cat# C15200229-100 Lot# 002). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
TurboCas: A method for locus-specific labeling of genomic regions and isolating their associated protein interactome
Bercin K Cenik et al.
Regulation of gene expression during development and stress response requires the concerted action of transcription factors and chromatin-binding proteins. Because this process is cell-type specific and varies with cellular conditions, mapping of chromatin factors at individual regulatory loci is crucial for underst... |
Genome-wide CRISPR screen identifies protein pathways modulating tauprotein levels in neurons
Sanchez C. G. et al.
Aggregates of hyperphosphorylated tau protein are a pathological hallmark of more than 20 distinct neurodegenerative diseases, including Alzheimer’s disease, progressive supranuclear palsy, and frontotemporal dementia. While the exact mechanism of tau aggregation is unknown, the accumulation of aggregates corr... |
Transgenic mice for in vivo epigenome editing with CRISPR-based systems
Gemberling, M. et al.
The discovery, characterization, and adaptation of the RNA-guided clustered regularly interspersed short palindromic repeat (CRISPR)-Cas9 system has greatly increased the ease with which genome and epigenome editing can be performed. Fusion of chromatin-modifying domains to the nuclease-deactivated form of Cas9 (dCa... |
Circuit-specific hippocampal ΔFosB underlies resilience to
stress-induced social avoidance.
Eagle, Andrew L and Manning, Claire E and Williams, Elizabeth S and Bastle,
Ryan M and Gajewski, Paula A and Garrison, Amber and Wirtz, Alexis J and
Akguen, Seda and Brandel-Ankrapp, Katie and Endege, Wilson and Boyce,
Frederick M and Ohnishi, Yoshinori N
Chronic stress is a key risk factor for mood disorders like depression, but
the stress-induced changes in brain circuit function and gene expression
underlying depression symptoms are not completely understood, hindering
development of novel treatments. Because of its projections to brain
regions regulating reward a... |
Endogenous retroviruses are a source of enhancers with oncogenic potential in acute myeloid leukaemia
/
Acute myeloid leukemia (AML) is a highly aggressive hematopoietic malignancy, defined by a series of genetic and epigenetic alterations, which result in deregulation of transcriptional networks. One understudied but important source of transcriptional regulators are transposable elements (TEs), which are widespread ... |
Improved double-nicking strategies for COL7A1 editing by homologous recombination
Kocher Thomas, Wagner Roland N., Klausegger Alfred, Guttmann-Gruber Christina, Hainzl Stefan, Bauer Johann W., Reichelt Julia, Koller Ulrich
Current gene editing approaches for treatment of recessive dystrophic epidermolysis bullosa (RDEB), an inherited, severe form of blistering skin disease, suffer from low efficiencies and safety concerns that complicate implementation in clinical settings. We present a strategy for efficient and precise repair of RDE... |
Improved Double-Nicking Strategies for COL7A1-Editing by Homologous Recombination.
Kocher T, Wagner RN, Klausegger A, Guttmann-Gruber C, Hainzl S, Bauer JW, Reichelt J, Koller U
Current gene-editing approaches for treatment of recessive dystrophic epidermolysis bullosa (RDEB), an inherited, severe form of blistering skin disease, suffer from low efficiencies and safety concerns that complicate implementation in clinical settings. We present a strategy for efficient and precise repair of RDE... |
Essential Gene Profiles for Human Pluripotent Stem Cells Identify Uncharacterized Genes and Substrate Dependencies.
Mair B, Tomic J, Masud SN, Tonge P, Weiss A, Usaj M, Tong AHY, Kwan JJ, Brown KR, Titus E, Atkins M, Chan KSK, Munsie L, Habsid A, Han H, Kennedy M, Cohen B, Keller G, Moffat J
Human pluripotent stem cells (hPSCs) provide an invaluable tool for modeling diseases and hold promise for regenerative medicine. For understanding pluripotency and lineage differentiation mechanisms, a critical first step involves systematically cataloging essential genes (EGs) that are indispensable for hPSC fitne... |
RNA-Based dCas9–VP64 System Improves the Viability of Cryopreserved Mammalian Cells
Hu Yong, Li Lei, Yu Yin, Huang Haishui, Uygun Basak E., Yarmush Martin L.
Regenerative therapies require availability of an abundant healthy cell source which can be achieved by e±cient cryopreservation techniques. Here, we established a novel approach for improved cell cryopreservation using an mRNA-based dCas9-VP64 gene activation system for transient, yet highly e±cient e... |
CRISPR-Mediated Gene Targeting of Human Induced Pluripotent Stem Cells
Susan M. Byrne, George M. Church
CRISPR/Cas9 nuclease systems can create double-stranded DNA breaks at specific sequences to efficiently and precisely disrupt, excise, mutate, insert, or replace genes. However, human embryonic stem cells and induced pluripotent stem cells (iPSCs) are more difficult to transfect and less resilient to DNA damage than... |