How to properly cite our product/service in your work We strongly recommend using this: RRBS service (Reduced Representation Bisulfite Sequencing) (Hologic Diagenode Cat# G02020000). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Mission SpaceX CRS-19 RRRM-1 space flight induced skin genomic plasticity via an epigenetic trigger
Kanhaiya Singh et al.
Highlights
Exposure to space environment causes genome-wide adaptive epigenetic changes
Space-exposure adaptive genome-wide changes are only seen in select “responder” mice
In space, genome-wide epigenetic changes mark induction of genomic plasticity
Genome-wide hypomethylation in... |
Long-term effects of myo-inositol on traumatic brain injury: Epigenomic and transcriptomic studies
Oganezovi N. et al.
Background and purpose
Traumatic brain injury (TBI) and its consequences remain great challenges for neurology. Consequences of TBI are associated with various alterations in the brain but little is known about long-term changes of epigenetic DNA methylation patterns. Moreover, nothing is known about potential trea... |
Gestational Caloric Restriction Alters Adipose Tissue Methylome and Offspring’s Metabolic Profile in a Swine Model
Mas-Pares B. et al.
Limited nutrient supply to the fetus results in physiologic and metabolic adaptations that have unfavorable consequences in the offspring. In a swine animal model, we aimed to study the effects of gestational caloric restriction and early postnatal metformin administration on offspring’s adipose tissue epigene... |
Diagnostic Algorithm to Subclassify Atypical Spitzoid Tumors in Low and High Risk According to Their Methylation Status
Gonzales-Munoz J.F. et al.
Current diagnostic algorithms are insufficient for the optimal clinical and therapeutic management of cutaneous spitzoid tumors, particularly atypical spitzoid tumors (AST). Therefore, it is crucial to identify new markers that allow for reliable and reproducible diagnostic assessment and can also be used as a predi... |
Sperm DNA methylation is predominantly stable in mice offspring bornafter transplantation of long-term cultured spermatogonial stem cells.
Serrano J. B.et al.
BACKGROUND: Spermatogonial stem cell transplantation (SSCT) is proposed as a fertility therapy for childhood cancer survivors. SSCT starts with cryopreserving a testicular biopsy prior to gonadotoxic treatments such as cancer treatments. When the childhood cancer survivor reaches adulthood and desires biological chi... |
Myelodysplastic Syndrome associated TET2 mutations affect NK cellfunction and genome methylation.
Boy M. et al.
Myelodysplastic syndromes (MDS) are clonal hematopoietic disorders, representing high risk of progression to acute myeloid leukaemia, and frequently associated to somatic mutations, notably in the epigenetic regulator TET2. Natural Killer (NK) cells play a role in the anti-leukemic immune response via their cytolyti... |
Epigenetics and stroke: role of DNA methylation and effect of aging onblood-brain barrier recovery.
Phillips C. et al.
Incomplete recovery of blood-brain barrier (BBB) function contributes to stroke outcomes. How the BBB recovers after stroke remains largely unknown. Emerging evidence suggests that epigenetic factors play a significant role in regulating post-stroke BBB recovery. This study aimed to evaluate the epigenetic and trans... |
Altered DNA methylation in estrogen-responsive repetitive sequences ofspermatozoa of infertile men with shortened anogenital distance.
Stenz L. et al.
BACKGROUND: It has been suggested that antenatal exposure to environmental endocrine disruptors is responsible for adverse trends in male reproductive health, including male infertility, impaired semen quality, cryptorchidism and testicular cancer, a condition known as testicular dysgenesis syndrome. Anogenital dist... |
Epigenetic Alterations of Repeated Relapses in Patient-matchedChildhood Ependymomas.
Zhao Sibo et al.
Recurrence is frequent in pediatric ependymoma (EPN). Our longitudinal integrated analysis of 30 patient-matched repeated relapses (3.67 ± 1.76 times) over 13 years (5.8 ± 3.8) reveals stable molecular subtypes (RELA and PFA) and convergent DNA methylation reprogramming duri... |
DNA methylation may affect beef tenderness through signal transduction inBos indicus.
de Souza M. M. et al.
BACKGROUND: Beef tenderness is a complex trait of economic importance for the beef industry. Understanding the epigenetic mechanisms underlying this trait may help improve the accuracy of breeding programs. However, little is known about epigenetic effects on Bos taurus muscle and their implications in tenderness, a... |
Genome-Wide Epigenomic Analyses in Patients With Nociceptive and Neuropathic Chronic Pain Subtypes Reveals Alterations in Methylation of Genes Involved in the Neuro-Musculoskeletal System
Stenz et al
Nociceptive pain involves the activation of nociceptors without damage to the nervous system, whereas neuropathic pain is related to an alteration in the central or peripheral nervous system. Chronic pain itself and the transition from acute to chronic pain may be epigenetically controlled. In this cross-sectional s... |
Genome-wide epigenomic analyses in patients with nociceptive andneuropathic chronic pain subtypes reveals alterations in methylation ofgenes involved in the neuro-musculoskeletal system.
Stenz Ludwig et al.
Nociceptive pain involves the activation of nociceptors without damage to the nervous system, whereas neuropathic pain is related to an alteration in the central or peripheral nervous system. Chronic pain itself and the transition from acute to chronic pain may be epigenetically controlled. In this cross-sectional s... |
Environmental enrichment preserves a young DNA methylation landscape in the aged mouse hippocampus
Sara Zocher, Rupert W. Overall, Mathias Lesche, Andreas Dahl & Gerd Kempermann
The decline of brain function during aging is associated with epigenetic changes, including DNA methylation. Lifestyle interventions can improve brain function during aging, but their influence on age-related epigenetic changes is unknown. Using genome-wide DNA methylation sequencing, we here show that experiencing ... |
Pathophysiological adaptations of resistance arteries in rat offspringexposed in utero to maternal obesity is associated with sex-specificepigenetic alterations.
Payen Cyrielle et al.
BACKGROUND/OBJECTIVES: Maternal obesity impacts vascular functions linked to metabolic disorders in offspring, leading to cardiovascular diseases during adulthood. Even if the relation between prenatal conditioning of cardiovascular diseases by maternal obesity and vascular function begins to be documented, little i... |
Muscle allele-specific expression QTLs may affect meat quality traitsin Bos indicus.
Bruscadin J.J. et al.
Single nucleotide polymorphisms (SNPs) located in transcript sequences showing allele-specific expression (ASE SNPs) were previously identified in the Longissimus thoracis muscle of a Nelore (Bos indicus) population consisting of 190 steers. Given that the allele-specific expression pattern may result from cis-regul... |
IGFBP2 protects against pulmonary fibrosis through inhibiting P21-mediated senescence
Chiahsuan, C. et al.
Accumulation of senescent cells contributes to age related diseases including idiopathic pulmonary fibrosis (IPF). Insulin-like growth factor binding proteins (IGFBPs) are evolutionarily conserved proteins that play a vital role in many biological processes. Overall, little is known about the functions of IGFBP2 in ... |
Perturbed DNA methylation by sustained overexpression of Gadd45b induces chromatin disorganization, DNA strand breaks and dopaminergic neurondeath in mice
Ravel-Godreuil, C. et al.
Heterochromatin disorganization is a key hallmark of aging and DNA methylation state is currently the main molecular predictor of chronological age. The most frequent neurodegenerative diseases like Parkinson disease and Alzheimer’s disease are age-related but how the aging process and chromatin alterations ar... |
Developmental cannabidiol exposure increases anxiety and modifiesgenome-wide brain DNA methylation in adult female mice.
Wanner N. M. et al.
BACKGROUND: Use of cannabidiol (CBD), the primary non-psychoactive compound found in cannabis, has recently risen dramatically, while relatively little is known about the underlying molecular mechanisms of its effects. Previous work indicates that direct CBD exposure strongly impacts the brain, with anxiolytic, anti... |
Hepatic transcriptome and DNA methylation patterns following perinataland chronic BPS exposure in male mice.
Brulport A. et al.
BACKGROUND: Bisphenol S (BPS) is a common bisphenol A (BPA) substitute, since BPA is virtually banned worldwide. However, BPS and BPA have both endocrine disrupting properties. Their effects appear mostly in adulthood following perinatal exposures. The objective of the present study was to investigate the impact of ... |
Integrative Analysis of Glucometabolic Traits, Adipose Tissue DNA Methylation and Gene Expression Identifies Epigenetic Regulatory Mechanisms of Insulin Resistance and Obesity in African Americans
Neeraj K. Sharma, Mary E. Comeau, Dennis Montoya, Matteo Pellegrini, Timothy D. Howard, Carl D. Langefeld, Swapan K. Das
Decline in insulin sensitivity due to dysfunction of adipose tissue (AT) is one of the earliest pathogenic events in Type 2 Diabetes. We hypothesize that differential DNA methylation (DNAm) controls insulin sensitivity and obesity by modulating transcript expression in AT. Integrating AT DNAm profiles with transcrip... |
Chronic cannabidiol alters genome-wide DNA methylation in adult mouse hippocampus: epigenetic implications for psychiatric disease.
Wanner NM, Colwell M, Drown C, Faulk C
Cannabidiol (CBD) is the primary non-psychoactive compound found in cannabis (Cannabis sativa) and an increasingly popular dietary supplement as a result of widespread availability of CBD-containing products. CBD is FDA-approved for the treatment of epilepsy and exhibits anxiolytic, antipsychotic, prosocial, and oth... |
LY75 Ablation Mediates Mesenchymal-Epithelial Transition (MET) in Epithelial Ovarian Cancer (EOC) Cells Associated with DNA Methylation Alterations and Suppression of the Wnt/β-Catenin Pathway.
Mehdi S, Bachvarova M, Scott-Boyer MP, Droit A, Bachvarov D
Growing evidence demonstrates that epithelial-mesenchymal transition (EMT) plays an important role in epithelial ovarian cancer (EOC) progression and spreading; however, its molecular mechanisms remain poorly defined. We have previously shown that the antigen receptor LY75 can modulate EOC cell phenotype and metasta... |
Obesogen effect of bisphenol S alters mRNA expression and DNA methylation profiling in male mouse liver
Brulport Axelle, Vaiman Daniel, Chagnon Marie-Christine, Le Corre Ludovic
Environmental pollution is increasingly considered an important factor involved in the obesity incidence. Endocrine disruptors (EDs) are important actors in the concept of DOHaD (Developmental Origins of Health and Disease), where epigenetic mechanisms play crucial roles. Bisphenol A (BPA), a monomer used in the man... |
Demethylation of ITGAV accelerates osteogenic differentiation in a blast-induced heterotopic ossification in vitro cell culture model.
Logan NJ, Camman M, Williams G, Higgins CA
Trauma-induced heterotopic ossification is an intriguing phenomenon involving the inappropriate ossification of soft tissues within the body such as the muscle and ligaments. This inappropriate formation of bone is highly prevalent in those affected by blast injuries. Here, we developed a simplified cell culture mod... |
Paternal sepsis induces alterations of the sperm methylome and dampens offspring immune responses—an animal study
Katharina Bomans, Judith Schenz, Sandra Tamulyte, Dominik Schaack, Markus Alexander Weigand and Florian Uhle
Abstract
Background
Sepsis represents the utmost severe consequence of infection, involving a dysregulated and self-damaging immune response of the host. While different environmental exposures like chronic stress or malnutrition have been well described to reprogram the germline and subsequently offspring a... |
DNMT3B overexpression contributes to aberrant DNA methylation and MYC-driven tumor maintenance in T-ALL and Burkitt’s lymphoma
Poole et al.
Aberrant DNA methylation is a hallmark of cancer. However, our understanding of how tumor cell-specific DNA methylation patterns are established and maintained is limited. Here, we report that in T-cell acute lymphoblastic leukemia (T-ALL) and Burkitt’s lymphoma the MYC oncogene causes overexpression of DNA me... |
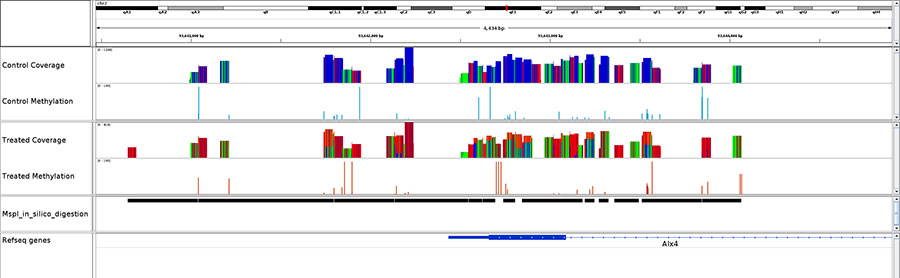
Diagenode® Premium RRBS technology: cost-effective DNA methylation mapping with superior coverage
Anne-Clémence Veillard, Paul Datlinger, Miklos Laczik, Sharon Squazzo & Christoph Bock
Reduced representation bisulfite sequencing (RRBS) enables genome-scale DNA methylation analysis in any vertebrate species. The assay benefits from the practical advantages of bisulfite sequencing while avoiding the cost of whole-genome sequencing. The Diagenode Premium RRBS kit makes this technology widely availabl... |