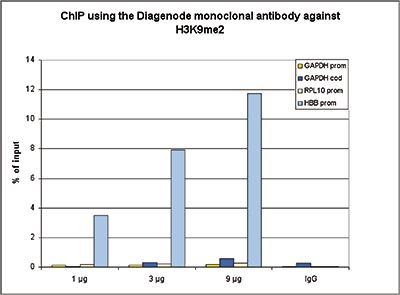
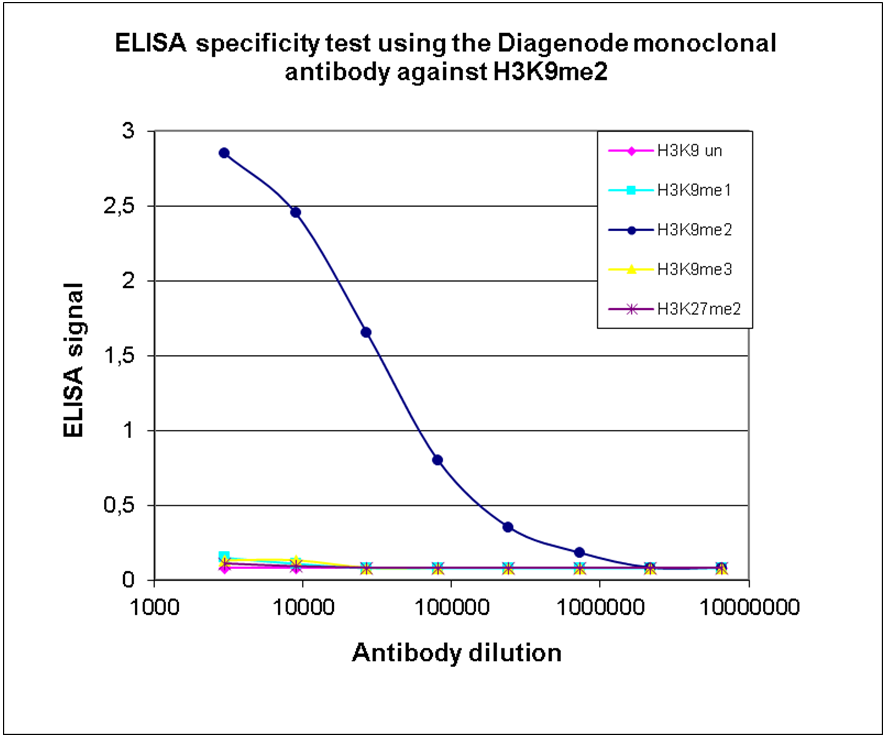
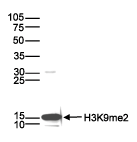
How to properly cite our product/service in your work We strongly recommend using this: H3K9me2 Antibody (sample size) (Hologic Diagenode Cat# C15200154-10 Lot# 001-013). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Histone lysine demethylase inhibition reprograms prostate cancermetabolism and mechanics.
Chianese Ugo and Papulino Chiara and Passaro Eugenia andEvers Tom Mj and Babaei Mehrad and Toraldo Antonella andDe Marchi Tommaso and Niméus Emma and Carafa Vincenzo andNicoletti Maria Maddalena and Del Gaudio Nunzio andIaccarino Nunzia an
OBJECTIVE: Aberrant activity of androgen receptor (AR) is the primary cause underlying development and progression of prostate cancer (PCa) and castration-resistant PCa (CRPC). Androgen signaling regulates gene transcription and lipid metabolism, facilitating tumor growth and therapy resistance in early and advanced... |
Histone post-translational modifications in Silene latifolia X and Y chromosomes suggest a mammal-like dosage compensation system
Luis Rodríguez Lorenzo José, Hubinský Marcel, Vyskot Boris, Hobza Roman
Silene latifolia is a model organism to study evolutionary young heteromorphic sex chromosome evolution in plants. Previous research indicates a Y-allele gene degeneration and a dosage compensation system already operating. Here, we propose an epigenetic approach based on analysis of several histone post-translation... |
The histone methyltransferase DOT1L is required for proper DNA damage response, DNA repair, and modulates chemotherapy responsiveness.
Kari V, Raul SK, Henck JM, Kitz J, Kramer F, Kosinsky RL, Übelmesser N, Mansour WY, Eggert J, Spitzner M, Najafova Z, Bastians H, Grade M, Gaedcke J, Wegwitz F, Johnsen SA
BACKGROUND: Disruptor of telomeric silencing 1-like (DOT1L) is a non-SET domain containing methyltransferase known to catalyze mono-, di-, and tri-methylation of histone 3 on lysine 79 (H3K79me). DOT1L-mediated H3K79me has been implicated in chromatin-associated functions including gene transcription, heterochromati... |
Role of Annexin gene and its regulation during zebrafish caudal fin regeneration
Saxena S, Purushothaman S, Meghah V, Bhatti B, Poruri A, Meena Lakshmi MG, Sarath Babu N, Murthy CL, Mandal KK, Kumar A, Idris MM
The molecular mechanism of epimorphic regeneration is elusive due to its complexity and limitation in mammals. Epigenetic regulatory mechanisms play a crucial role in development and regeneration. This investigation attempted to reveal the role of epigenetic regulatory mechanisms, such as histone H3 and H4 lysine ac... |
Independent Mechanisms Target SMCHD1 to Trimethylated Histone H3 Lysine 9-Modified Chromatin and the Inactive X Chromosome
Brideau NJ, Coker H, Gendrel AV, Siebert CA, Bezstarosti K, Demmers J, Poot RA, Nesterova TB, Brockdorff N
The chromosomal protein SMCHD1 plays an important role in epigenetic silencing at diverse loci, including the inactive X chromosome, imprinted genes, and the facioscapulohumeral muscular dystrophy locus. Although homology with canonical SMC family proteins suggests a role in chromosome organization, the mechanisms u... |
A gene expression signature identifying transient DNMT1 depletion as a causal factor of cancer-germline gene activation in melanoma
Charles De Smet
Many human tumors show aberrant activation of a group of germline-specific genes, termed cancer-germline (CG) genes, several of which appear to exert oncogenic functions. Although activation of CG genes in tumors has been linked to promoter DNA demethylation, the mechanisms underlying this epigenetic alteration rema... |
Epigenetic hierarchy within the MAGEA1 cancer-germline gene: promoter DNA methylation dictates local histone modifications.
Cannuyer J, Loriot A, Parvizi GK, De Smet C
Gene MAGEA1 belongs to a group of human germline-specific genes that rely on DNA methylation for repression in somatic tissues. Many of these genes, termed cancer-germline (CG) genes, become demethylated and activated in a wide variety of tumors, where they encode tumor-specific antigens. The process leading to DNA ... |