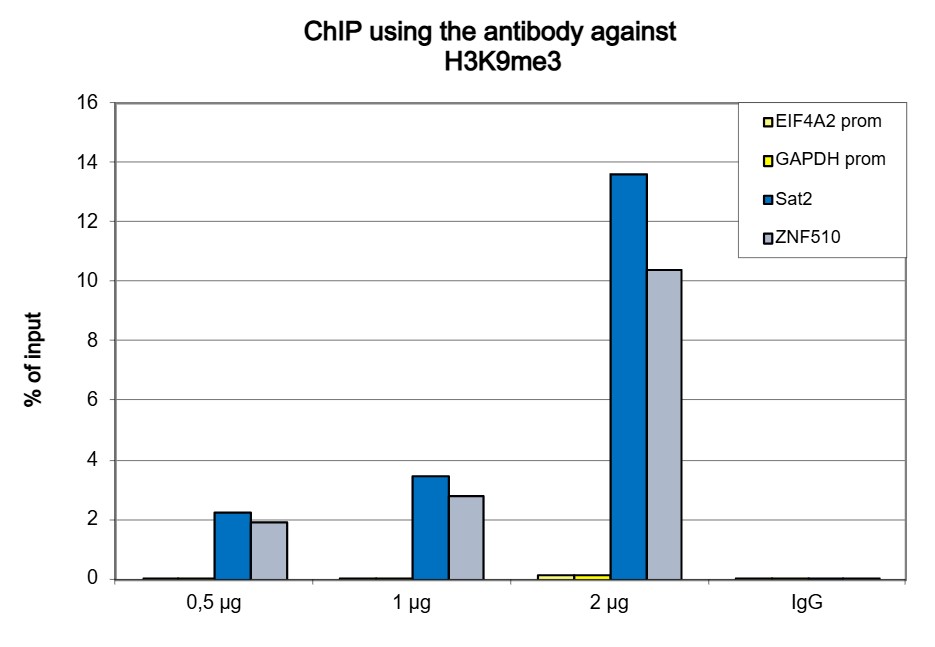
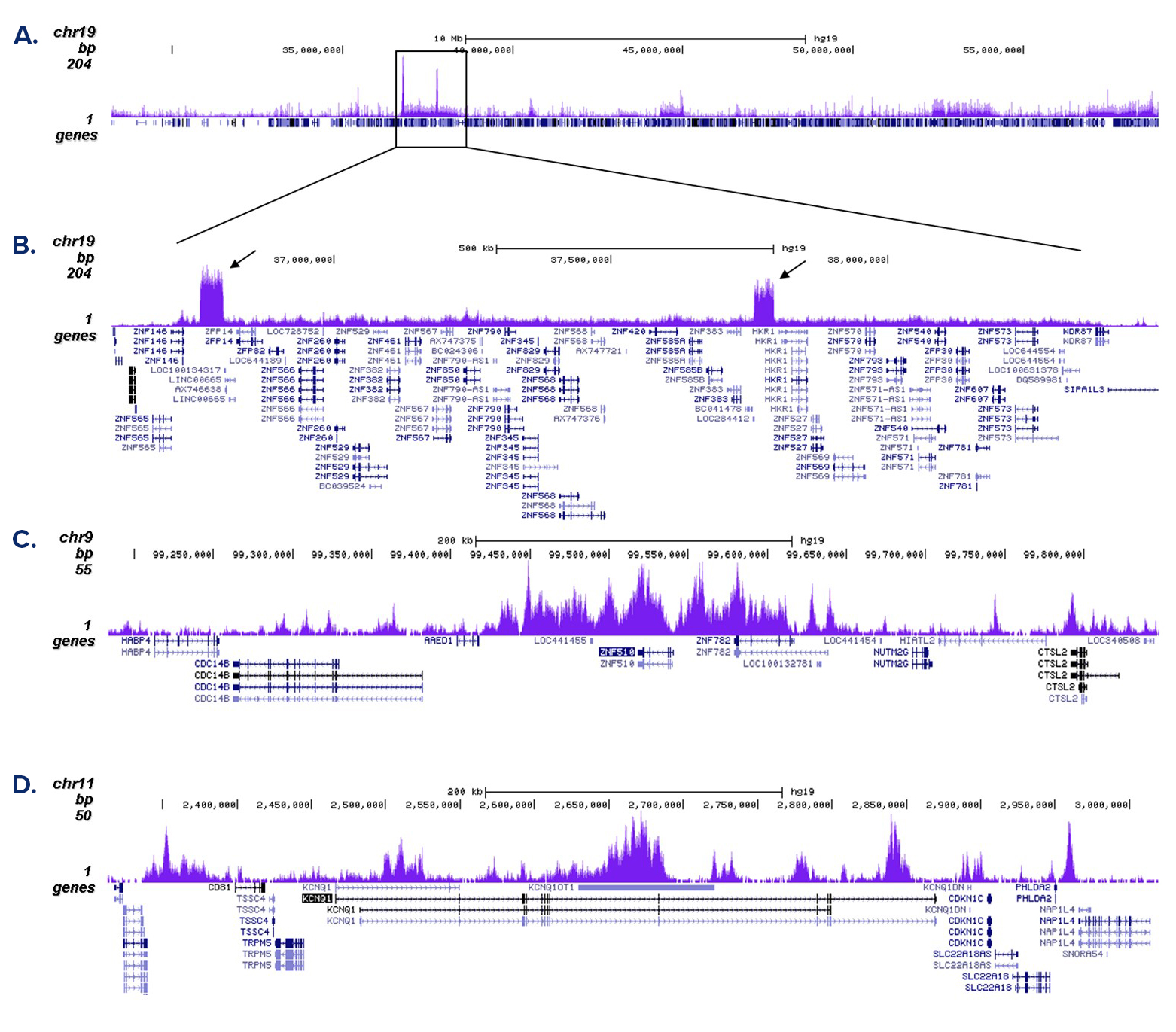
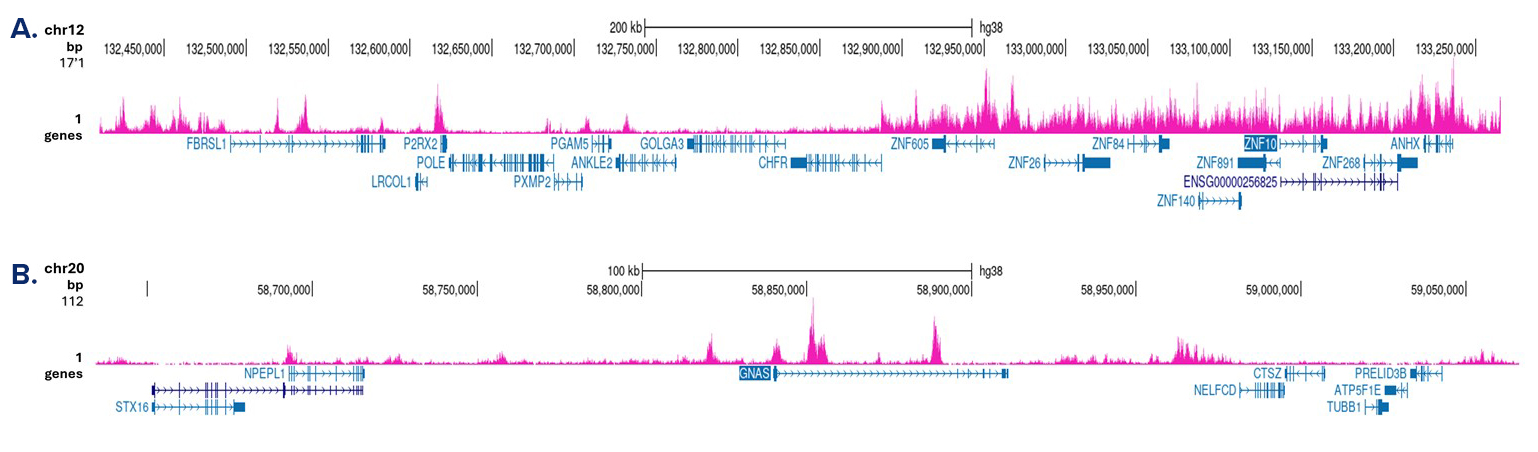
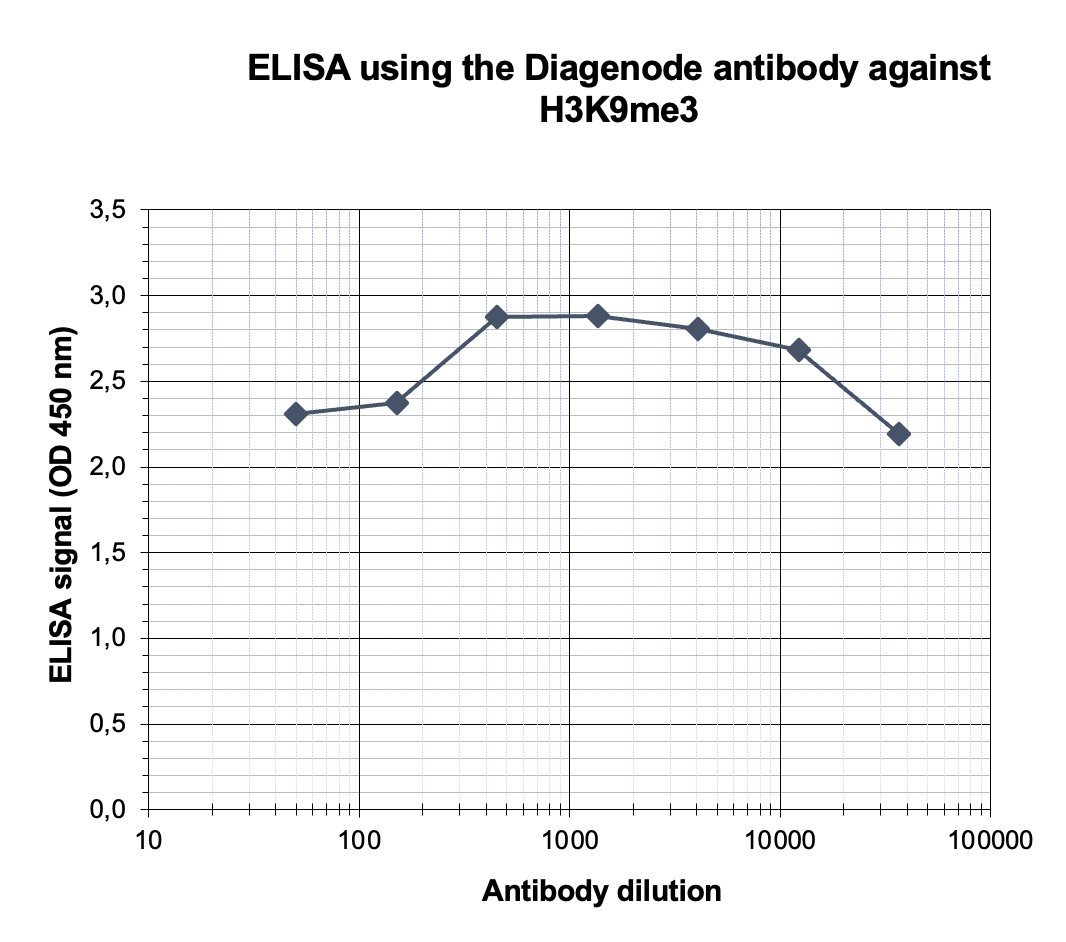
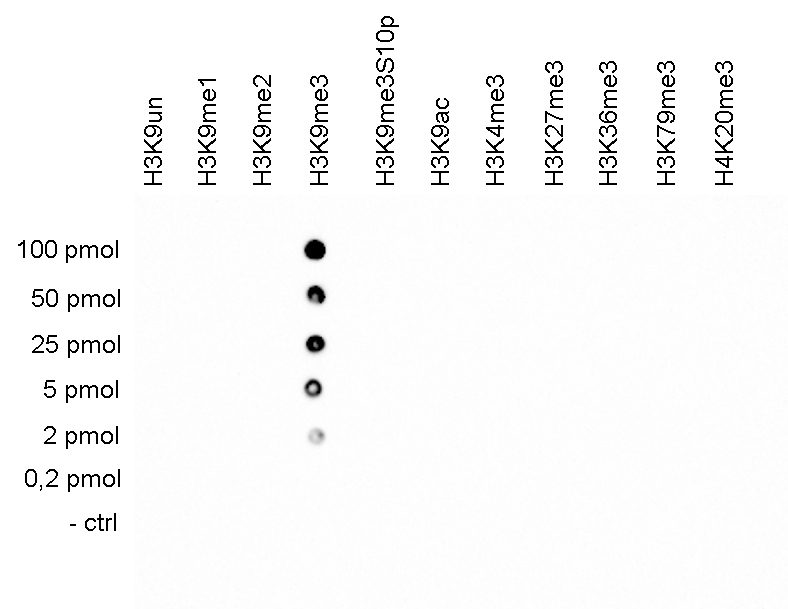
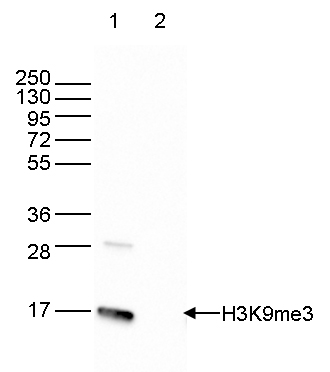
How to properly cite our product/service in your work We strongly recommend using this: H3K9me3 Antibody (Hologic Diagenode Cat# C15410056 Lot# A2810P). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Multimodal epigenetic and enhancer network remodeling shape the transcriptional landscape of human beige adipocytes
Hazell Pickering, Sarah et al.
Epigenetic regulation is a key determinant of adipocyte fate, driving the differentiation toward white or thermogenic beige phenotypes in response to environmental cues. To dissect the mechanisms orchestrating this plasticity in human adipocytes, we conducted an integrative analysis of transcriptomic, epigenomic... |
Multimodal epigenetic and enhancer network remodeling shape the transcriptional landscape of human beige adipocytes
Hazell Pickering, Sarah et al.
Abstract
Epigenetic regulation is a key determinant of adipocyte fate, driving the differentiation toward white or thermogenic beige phenotypes in response to environmental cues. To dissect the mechanisms orchestrating this plasticity in human adipocytes, we conducted an integrative analysis of transcriptomic, epig... |
Refeeding with different levels of dietary carbohydrate modulates epigenetic stability through global DNA (de)methylation and histone modifications in juvenile and adult Nile tilapia (Oreochromis niloticus)
Thongchaitriwat, Sirijanya et al.
The Nile tilapia (Oreochromis niloticus) exhibits a strong metabolic response to dietary carbohydrates (CHO). Short-term refeeding after fasting with a high-carbohydrate (HC) diet has been shown to modulate CHO metabolism, but the role of epigenetic regulation in this response remains unclear. This study investiga... |
Analog epigenetic memory revealed by targeted chromatin editing
Palacios, Sebastian et al.
Cells store information by means of chromatin modifications that persist through cell divisions and can hold gene expression silenced over generations. However, how these modifications may maintain other gene expression states has remained unclear. This study shows that chromatin modifications can maintain a wide ... |
Ectopic expression of DNMT3L in human trophoblast stem cells restores features of the placental methylome
Lea, Georgia et al.
The placental DNA methylation landscape is unique, with widespread partially methylated domains (PMDs). The placental "methylome" is conserved across mammals, a shared feature of many cancers, and extensively studied for links with pregnancy complications. Human trophoblast stem cells (hTSCs) offer exciting potent... |
SMYD3 represses tumor-intrinsic interferon response in HPV-negativesquamous cell carcinoma of the head and neck.
Nigam N. et al.
Cancers often display immune escape, but the mechanisms are incompletely understood. Herein, we identify SMYD3 as a mediator of immune escape in human papilloma virus (HPV)-negative head and neck squamous cell carcinoma (HNSCC), an aggressive disease with poor response to immunotherapy with pembrolizumab. SMYD3 depl... |
RUNX1 colludes with NOTCH1 to reprogram chromatin in T-cell acutelymphoblastic leukemia
Islam R. et al.
Runt-related transcription factor 1 (RUNX1) is oncogenic in diverse types of leukemia and epithelial cancers where its expression is associated with poor prognosis. Current models suggest that RUNX1 cooperates with other oncogenic factors (e.g., NOTCH1, TAL1) to drive the expression of proto-oncogenes in T cell... |
Gene Regulatory Interactions at Lamina-Associated Domains
Madsen-Østerbye J. et al.
The nuclear lamina provides a repressive chromatin environment at the nuclear periphery. However, whereas most genes in lamina-associated domains (LADs) are inactive, over ten percent reside in local euchromatic contexts and are expressed. How these genes are regulated and whether they are able to interact with regu... |
Determinants of heritable gene silencing for KRAB-dCas9 + DNMT3and Ezh2-dCas9 + DNMT3 hit-and-run epigenome editing.
O'Geen H.et al.
Precision epigenome editing has gained significant attention as a method to modulate gene expression without altering genetic information. However, a major limiting factor has been that the gene expression changes are often transient, unlike the life-long epigenetic changes that occur frequently in nature. Here, we ... |
The prolyl-isomerase PIN1 is essential for nuclear Lamin-Bstructure and function and protects heterochromatin under mechanicalstress.
Napoletano Francesco et al.
Chromatin organization plays a crucial role in tissue homeostasis. Heterochromatin relaxation and consequent unscheduled mobilization of transposable elements (TEs) are emerging as key contributors of aging and aging-related pathologies, including Alzheimer's disease (AD) and cancer. However, the mechanisms governin... |
Dynamic association of the H3K64 trimethylation mark with genes encodingexported proteins in Plasmodium falciparum.
Jabeena, C A et al.
Epigenetic modifications have emerged as critical regulators of virulence genes and stage-specific gene expression in Plasmodium falciparum. However, the specific roles of histone core epigenetic modifications in regulating the stage-specific gene expression are not well understood. In this study, we report an uncon... |
Restricted nucleation and piRNA-mediated establishment of heterochromatinduring embryogenesis in Drosophila miranda
Wei, K. et al.
Heterochromatin is a key architectural feature of eukaryotic genomes, crucial for silencing of repetitive elements and maintaining genome stability. Heterochromatin shows stereotypical enrichment patterns around centromeres and repetitive sequences, but the molecular details of how heterochromatin is established dur... |
Epigenetic, transcriptional and phenotypic responses in Daphnia magna exposed to low-level ionizing radiation
Thaulow Jens, Song You, Lindeman Leif C., Kamstra Jorke H., Lee YeonKyeong, Xie Li, Aleström Peter, Salbu Brit, Tollefsen Knut Erik
Ionizing radiation is known to induce oxidative stress and DNA damage as well as epigenetic effects in aquatic organisms. Epigenetic changes can be part of the adaptive responses to protect organisms from radiation-induced damage, or act as drivers of toxicity pathways leading to adverse effects. To investigate the ... |
The 20S proteasome activator PA28γ controls the compaction of chromatin
Didier Fesquet, David Llères, Cristina Viganò, Francisca Méchali, Séverine Boulon, Robert Feil, Olivier Coux, Catherine Bonne-Andrea, Véronique Baldin
The nuclear PA28γ is known to activate the 20S proteasome, but its precise cellular functions remains unclear. Here, we identify PA28γ as a key factor that structures heterochromatin. We find that in human cells, a fraction of PA28γ-20S proteasome complexes localizes within HP1-linked heterochromat... |
Histone post-translational modifications in Silene latifolia X and Y chromosomes suggest a mammal-like dosage compensation system
Luis Rodríguez Lorenzo José, Hubinský Marcel, Vyskot Boris, Hobza Roman
Silene latifolia is a model organism to study evolutionary young heteromorphic sex chromosome evolution in plants. Previous research indicates a Y-allele gene degeneration and a dosage compensation system already operating. Here, we propose an epigenetic approach based on analysis of several histone post-translation... |
SETDB2 promoted breast cancer stem cell maintenance by interaction with and stabilization of ΔNp63α protein
Ying Liu, Fei Xie, Jialun Li, Jianpeng Xiao, Jie Wang, Zhaolin Mei, Hongjia Fan, Huan Fang, Sha Li, Qiuju Wu, Lin Yuan, Cuicui Liu, You Peng, Weiwei Zhao, Lulu Wang, Jiemin Wong, Jing Li, Jing Feng
The histone H3K9 methyltransferase SETDB2 is involved in cell cycle dysregulation in acute leukemia and has oncogenic roles in gastric cancer. In our study, we found that SETDB2 plays essential roles in breast cancer stem cell maintenance. Depleted SETDB2 significantly decreased the breast cancer stem cell populatio... |
Transit amplifying cells coordinate mouse incisor mesenchymal stem cell activation.
Walker JV, Zhuang H, Singer D, Illsley CS, Kok WL, Sivaraj KK, Gao Y, Bolton C, Liu Y, Zhao M, Grayson PRC, Wang S, Karbanová J, Lee T, Ardu S, Lai Q, Liu J, Kassem M, Chen S, Yang K, Bai Y, Tredwin C, Zambon AC, Corbeil D, Adams R, Abdallah BM, Hu B
Stem cells (SCs) receive inductive cues from the surrounding microenvironment and cells. Limited molecular evidence has connected tissue-specific mesenchymal stem cells (MSCs) with mesenchymal transit amplifying cells (MTACs). Using mouse incisor as the model, we discover a population of MSCs neibouring to the MTACs... |
Ezh2-dCas9 and KRAB-dCas9 enable engineering of epigenetic memory in a context-dependent manner.
O'Geen H, Bates SL, Carter SS, Nisson KA, Halmai J, Fink KD, Rhie SK, Farnham PJ, Segal DJ
BACKGROUND: Rewriting of the epigenome has risen as a promising alternative to gene editing for precision medicine. In nature, epigenetic silencing can result in complete attenuation of target gene expression over multiple mitotic divisions. However, persistent repression has been difficult to achieve in a predictab... |
Long-range interactions between topologically associating domains shape the four-dimensional genome during differentiation.
Paulsen J, Liyakat Ali TM, Nekrasov M, Delbarre E, Baudement MO, Kurscheid S, Tremethick D, Collas P
Genomic information is selectively used to direct spatial and temporal gene expression during differentiation. Interactions between topologically associating domains (TADs) and between chromatin and the nuclear lamina organize and position chromosomes in the nucleus. However, how these genomic organizers together sh... |
PML modulates H3.3 targeting to telomeric and centromeric repeats in mouse fibroblasts.
Spirkoski J, Shah A, Reiner AH, Collas P, Delbarre E
Targeted deposition of histone variant H3.3 into chromatin is paramount for proper regulation of chromatin integrity, particularly in heterochromatic regions including repeats. We have recently shown that the promyelocytic leukemia (PML) protein prevents H3.3 from being deposited in large heterochromatic PML-associa... |
Hominoid-Specific Transposable Elements and KZFPs Facilitate Human Embryonic Genome Activation and Control Transcription in Naive Human ESCs.
Pontis J, Planet E, Offner S, Turelli P, Duc J, Coudray A, Theunissen TW, Jaenisch R, Trono D
Expansion of transposable elements (TEs) coincides with evolutionary shifts in gene expression. TEs frequently harbor binding sites for transcriptional regulators, thus enabling coordinated genome-wide activation of species- and context-specific gene expression programs, but such regulation must be balanced against ... |
Gamma radiation induces locus specific changes to histone modification enrichment in zebrafish and Atlantic salmon.
Lindeman LC, Kamstra JH, Ballangby J, Hurem S, Martín LM, Brede DA, Teien HC, Oughton DH, Salbu B, Lyche JL, Aleström P
Ionizing radiation is a recognized genotoxic agent, however, little is known about the role of the functional form of DNA in these processes. Post translational modifications on histone proteins control the organization of chromatin and hence control transcriptional responses that ultimately affect the phenotype. Th... |
TIP60: an actor in acetylation of H3K4 and tumor development in breast cancer.
Judes G, Dubois L, Rifaï K, Idrissou M, Mishellany F, Pajon A, Besse S, Daures M, Degoul F, Bignon YJ, Penault-Llorca F, Bernard-Gallon D
AIM: The acetyltransferase TIP60 is reported to be downregulated in several cancers, in particular breast cancer, but the molecular mechanisms resulting from its alteration are still unclear. MATERIALS & METHODS: In breast tumors, H3K4ac enrichment and its link with TIP60 were evaluated by chromatin immunoprecip... |
PWWP2A binds distinct chromatin moieties and interacts with an MTA1-specific core NuRD complex.
Link S, Spitzer RMM, Sana M, Torrado M, Völker-Albert MC, Keilhauer EC, Burgold T, Pünzeler S, Low JKK, Lindström I, Nist A, Regnard C, Stiewe T, Hendrich B, Imhof A, Mann M, Mackay JP, Bartkuhn M, Hake SB
Chromatin structure and function is regulated by reader proteins recognizing histone modifications and/or histone variants. We recently identified that PWWP2A tightly binds to H2A.Z-containing nucleosomes and is involved in mitotic progression and cranial-facial development. Here, using in vitro assays, we show that... |
HIV-2/SIV viral protein X counteracts HUSH repressor complex.
Ghina Chougui, Soundasse Munir-Matloob, Roy Matkovic, Michaël M Martin, Marina Morel, Hichem Lahouassa, Marjorie Leduc, Bertha Cecilia Ramirez, Lucie Etienne and Florence Margottin-Goguet
To evade host immune defences, human immunodeficiency viruses 1 and 2 (HIV-1 and HIV-2) have evolved auxiliary proteins that target cell restriction factors. Viral protein X (Vpx) from the HIV-2/SIVsmm lineage enhances viral infection by antagonizing SAMHD1 (refs ), but this antagonism is not sufficient to explain a... |
Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes.
Lomberk G, Blum Y, Nicolle R, Nair A, Gaonkar KS, Marisa L, Mathison A, Sun Z, Yan H, Elarouci N, Armenoult L, Ayadi M, Ordog T, Lee JH, Oliver G, Klee E, Moutardier V, Gayet O, Bian B, Duconseil P, Gilabert M, Bigonnet M, Garcia S, Turrini O, Delpero JR,
Recent studies have offered ample insight into genome-wide expression patterns to define pancreatic ductal adenocarcinoma (PDAC) subtypes, although there remains a lack of knowledge regarding the underlying epigenomics of PDAC. Here we perform multi-parametric integrative analyses of chromatin immunoprecipitation-se... |
PRDM9 Methyltransferase Activity Is Essential for Meiotic DNA Double-Strand Break Formation at Its Binding Sites.
Diagouraga B, Clément JAJ, Duret L, Kadlec J, de Massy B, Baudat F
The programmed formation of hundreds of DNA double-strand breaks (DSBs) is essential for proper meiosis and fertility. In mice and humans, the location of these breaks is determined by the meiosis-specific protein PRDM9, through the DNA-binding specificity of its zinc-finger domain. PRDM9 also has methyltransferase ... |
DNA methylation of intragenic CpG islands depends on their transcriptional activity during differentiation and disease
Jeziorska D.M. et al.
The human genome contains ∼30,000 CpG islands (CGIs). While CGIs associated with promoters nearly always remain unmethylated, many of the ∼9,000 CGIs lying within gene bodies become methylated during development and differentiation. Both promoter and intragenic CGIs may also become abnormally methylated as a... |
Multivalent binding of PWWP2A to H2A.Z regulates mitosis and neural crest differentiation
Pünzeler S. et al.
Replacement of canonical histones with specialized histone variants promotes altering of chromatin structure and function. The essential histone variant H2A.Z affects various DNA-based processes via poorly understood mechanisms. Here, we determine the comprehensive interactome of H2A.Z and identify PWWP2A as a novel... |
Vitamin C induces specific demethylation of H3K9me2 in mouse embryonic stem cells via Kdm3a/b
Kevin T. Ebata, Kathryn Mesh, Shichong Liu, Misha Bilenky, Alexander Fekete, Michael G. Acker, Martin Hirst, Benjamin A. Garcia and Miguel Ramalho-Santos
Background
Histone methylation patterns regulate gene expression and are highly dynamic during development. The erasure of histone methylation is carried out by histone demethylase enzymes. We had previously shown that vitamin C enhances the activity of Tet enzymes in embryonic stem (ES) cells, leading to DNA... |
Microinjection of Antibodies Targeting the Lamin A/C Histone-Binding Site Blocks Mitotic Entry and Reveals Separate Chromatin Interactions with HP1, CenpB and PML.
Dixon C.R. et al.
Lamins form a scaffold lining the nucleus that binds chromatin and contributes to spatial genome organization; however, due to the many other functions of lamins, studies knocking out or altering the lamin polymer cannot clearly distinguish between direct and indirect effects. To overcome this obstacle, we specifica... |
Decoupling of DNA methylation and activity of intergenic LINE-1 promoters in colorectal cancer
Vafadar-Isfahani N. et al.
Hypomethylation of LINE-1 repeats in cancer has been proposed as the main mechanism behind their activation; this assumption, however, was based on findings from early studies that were biased toward young and transpositionally active elements. Here, we investigate the relationship between methylation of 2 intergeni... |
H3.Y discriminates between HIRA and DAXX chaperone complexes and reveals unexpected insights into human DAXX-H3.3-H4 binding and deposition requirements
Zink L.M. et al.
Histone chaperones prevent promiscuous histone interactions before chromatin assembly. They guarantee faithful deposition of canonical histones and functionally specialized histone variants into chromatin in a spatial- and temporally-restricted manner. Here, we identify the binding partners of the primate-specific a... |
Muscle catabolic capacities and global hepatic epigenome are modified in juvenile rainbow trout fed different vitamin levels at first feeding
Panserat S. et al.
Based on the concept of nutritional programming in mammals, we tested whether a short term hyper or hypo vitamin stimulus during first-feeding could induce long-lasting changes in nutrient metabolism in rainbow trout. Trout alevins received during the 4 first weeks of exogenous feeding a diet either without suppleme... |
Oestrogen receptor β regulates epigenetic patterns at specific genomic loci through interaction with thymine DNA glycosylase.
Liu Y, Duong W, Krawczyk C, Bretschneider N, Borbély G, Varshney M, Zinser C, Schär P, Rüegg J.
BACKGROUND:
DNA methylation is one way to encode epigenetic information and plays a crucial role in regulating gene expression during embryonic development. DNA methylation marks are established by the DNA methyltransferases and, recently, a mechanism for active DNA demethylation has emerged involving the ten-ele... |
Epigenetic priming of inflammatory response genes by high glucose in adipose progenitor cells
Rønningen T, Shah A, Reiner AH, Collas P, Moskaug JØ
Cellular metabolism confers wide-spread epigenetic modifications required for regulation of transcriptional networks that determine cellular states. Mesenchymal stromal cells are responsive to metabolic cues including circulating glucose levels and modulate inflammatory responses. We show here that long term exposur... |
Polycomb RING1A- and RING1B-dependent histone H2A monoubiquitylation at pericentromeric regions promotes S-phase progression
Bravo M, Nicolini F, Starowicz K, Barroso S, Calés C, Aguilera A, Vidal M
The functions of polycomb products extend beyond their well-known activity as transcriptional regulators to include genome duplication processes. Polycomb activities during DNA replication and DNA damage repair are unclear, particularly without induced replicative stress. We have used a cellular model of conditional... |
Erosion of X Chromosome Inactivation in Human Pluripotent Cells Initiates with XACT Coating and Depends on a Specific Heterochromatin Landscape.
Vallot C, Ouimette JF, Makhlouf M, Féraud O, Pontis J, Côme J, Martinat C, Bennaceur-Griscelli A, Lalande M, Rougeulle C
Human pluripotent stem cells (hPSCs) display extensive epigenetic instability, particularly on the X chromosome. In this study, we show that, in hPSCs, the inactive X chromosome has a specific heterochromatin landscape that predisposes it to erosion of X chromosome inactivation (XCI), a process that occurs spontaneo... |
Embryonic stem cell differentiation requires full length Chd1.
Piatti P, Lim CY, Nat R, Villunger A, Geley S, Shue YT, Soratroi C, Moser M, Lusser A
The modulation of chromatin dynamics by ATP-dependent chromatin remodeling factors has been recognized as an important mechanism to regulate the balancing of self-renewal and pluripotency in embryonic stem cells (ESCs). Here we have studied the effects of a partial deletion of the gene encoding the chromatin remodel... |
The histone demethylase enzyme KDM3A is a key estrogen receptor regulator in breast cancer.
Wade MA, Jones D, Wilson L, Stockley J, Coffey K, Robson CN, Gaughan L
Endocrine therapy has successfully been used to treat estrogen receptor (ER)-positive breast cancer, but this invariably fails with cancers becoming refractory to treatment. Emerging evidence has suggested that fluctuations in ER co-regulatory protein expression may facilitate resistance to therapy and be involved i... |
TRIM28 Represses Transcription of Endogenous Retroviruses in Neural Progenitor Cells.
Fasching L, Kapopoulou A, Sachdeva R, Petri R, Jönsson ME, Männe C, Turelli P, Jern P, Cammas F, Trono D, Jakobsson J
TRIM28 is a corepressor that mediates transcriptional silencing by establishing local heterochromatin. Here, we show that deletion of TRIM28 in neural progenitor cells (NPCs) results in high-level expression of two groups of endogenous retroviruses (ERVs): IAP1 and MMERVK10C. We find that NPCs use TRIM28-mediated hi... |
Cryosectioning the intestinal crypt-villus axis: an ex vivo method to study the dynamics of epigenetic modifications from stem cells to differentiated cells
Vincent A, Kazmierczak C, Duchêne B, Jonckheere N, Leteurtre E, Van Seuningen I
The intestinal epithelium is a particularly attractive biological adult model to study epigenetic mechanisms driving adult stem cell renewal and cell differentiation. Since epigenetic modifications are dynamic, we have developed an original ex vivo approach to study the expression and epigenetic profiles of key gene... |
Analysis of an artificial zinc finger epigenetic modulator: widespread binding but limited regulation.
Grimmer MR, Stolzenburg S, Ford E, Lister R, Blancafort P, Farnham PJ
Artificial transcription factors (ATFs) and genomic nucleases based on a DNA binding platform consisting of multiple zinc finger domains are currently being developed for clinical applications. However, no genome-wide investigations into their binding specificity have been performed. We have created six-finger ATFs ... |
The PML-associated protein DEK regulates the balance of H3.3 loading on chromatin and is important for telomere integrity.
Ivanauskiene K, Delbarre E, McGhie JD, Küntziger T, Wong LH, Collas P
Histone variant H3.3 is deposited in chromatin at active sites, telomeres, and pericentric heterochromatin by distinct chaperones, but the mechanisms of regulation and coordination of chaperone-mediated H3.3 loading remain largely unknown. We show here that the chromatin-associated oncoprotein DEK regulates differen... |
The specific alteration of histone methylation profiles by DZNep during early zebrafish development.
Ostrup O, Reiner AH, Aleström P, Collas P
Early embryo development constitutes a unique opportunity to study acquisition of epigenetic marks, including histone methylation. This study investigates the in vivo function and specificity of 3-deazaneplanocin A (DZNep), a promising anti-cancer drug that targets polycomb complex genes. One- to two-cell stage embr... |
KMTase Set7/9 is a critical regulator of E2F1 activity upon genotoxic stress.
Lezina L, Aksenova V, Ivanova T, Purmessur N, Antonov AV, Tentler D, Fedorova O, Garabadgiu AV, Talianidis I, Melino G, Barlev NA
During the recent years lysine methyltransferase Set7/9 ((Su(var)-3-9, Enhancer-of-Zeste, Trithorax) domain containing protein 7/9) has emerged as an important regulator of different transcription factors. In this study, we report a novel function for Set7/9 as a critical co-activator of E2 promoter-binding factor 1... |
The histone variant composition of centromeres is controlled by the pericentric heterochromatin state during the cell cycle.
Boyarchuk E, Filipescu D, Vassias I, Cantaloube S, Almouzni G
Correct chromosome segregation requires a unique chromatin environment at centromeres and in their vicinity. Here, we address how the deposition of canonical H2A and H2A.Z histone variants is controlled at pericentric heterochromatin (PHC). Whereas in euchromatin newly synthesized H2A and H2A.Z are deposited through... |
RNAi-Mediated Gene silencing in Zebrafish Triggered by Convergent Transcription.
Andrews OE, Cha DJ, Wei C, Patton JG
RNAi based strategies to induce gene silencing are commonly employed in numerous model organisms but have not been extensively used in zebrafish. We found that introduction of transgenes containing convergent transcription units in zebrafish embryos induced stable transcriptional gene silencing (TGS) in cis and tran... |
Transcriptional Derepression of the ERVWE1 Locus following Influenza A Virus Infection.
Li F, Nellåker C, Sabunciyan S, Yolken RH, Jones-Brando L, Johansson AS, Owe-Larsson B, Karlsson H
UNLABELLED: Syncytin-1, a fusogenic protein encoded by a human endogenous retrovirus of the W family (HERV-W) element (ERVWE1), is expressed in the syncytiotrophoblast layer of the placenta. This locus is transcriptionally repressed in adult tissues through promoter CpG methylation and suppressive histone modificati... |
HDAC inhibitor confers radiosensitivity to prostate stem-like cells.
Frame FM, Pellacani D, Collins AT, Simms MS, Mann VM, Jones G, Meuth M, Bristow RG, Maitland NJ
Background:Radiotherapy can be an effective treatment for prostate cancer, but radiorecurrent tumours do develop. Considering prostate cancer heterogeneity, we hypothesised that primitive stem-like cells may constitute the radiation-resistant fraction.Methods:Primary cultures were derived from patients undergoing re... |
Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes.
Lund E, Oldenburg AR, Delbarre E, Freberg CT, Duband-Goulet I, Eskeland R, Buendia B, Collas P
The nuclear lamina is implicated in the organization of the eukaryotic nucleus. Association of nuclear lamins with the genome occurs through large chromatin domains including mostly, but not exclusively, repressed genes. How lamin interactions with regulatory elements modulate gene expression in different cellular c... |
Long range epigenetic silencing is a trans-species mechanism that results in cancer specific deregulation by overriding the chromatin domains of normal cells.
Forn M, Muñoz M, Tauriello DV, Merlos-Suárez A, Rodilla V, Bigas A, Batlle E, Jordà M, Peinado MA
DNA methylation and chromatin remodeling are frequently implicated in the silencing of genes involved in carcinogenesis. Long Range Epigenetic Silencing (LRES) is a mechanism of gene inactivation that affects multiple contiguous CpG islands and has been described in different human cancer types. However, it is unkno... |
Transgene- and locus-dependent imprinting reveals allele-specific chromosome conformations.
Lonfat N, Montavon T, Jebb D, Tschopp P, Nguyen Huynh TH, Zakany J, Duboule D
When positioned into the integrin α-6 gene, an Hoxd9lacZ reporter transgene displayed parental imprinting in mouse embryos. While the expression from the paternal allele was comparable with patterns seen for the same transgene when present at the neighboring HoxD locus, almost no signal was scored at this integratio... |
Expression of a large LINE-1-driven antisense RNA is linked to epigenetic silencing of the metastasis suppressor gene TFPI-2 in cancer.
Cruickshanks HA, Vafadar-Isfahani N, Dunican DS, Lee A, Sproul D, Lund JN, Meehan RR, Tufarelli C
LINE-1 retrotransposons are abundant repetitive elements of viral origin, which in normal cells are kept quiescent through epigenetic mechanisms. Activation of LINE-1 occurs frequently in cancer and can enable LINE-1 mobilization but also has retrotransposition-independent consequences. We previously reported that i... |
Histone lysine trimethylation or acetylation can be modulated by phytoestrogen, estrogen or anti-HDAC in breast cancer cell lines.
Dagdemir A, Durif J, Ngollo M, Bignon YJ, Bernard-Gallon D
AIM: The isoflavones genistein, daidzein and equol (daidzein metabolite) have been reported to interact with epigenetic modifications, specifically hypermethylation of tumor suppressor genes. The objective of this study was to analyze and understand the mechanisms by which phytoestrogens act on chromatin in breast c... |
Epigenetic Regulation of Nestin Expression During Neurogenic Differentiation of Adipose Tissue Stem Cells.
Boulland JL, Mastrangelopoulou M, Boquest AC, Jakobsen R, Noer A, Glover JC, Collas P.
Adipose-tissue-derived stem cells (ASCs) have received considerable attention due to their easy access, expansion potential, and differentiation capacity. ASCs are believed to have the potential to differentiate into neurons. However, the mechanisms by which this may occur remain largely unknown. Here, we show that ... |
The histone demethylase Kdm3a is essential to progression through differentiation.
Herzog M, Josseaux E, Dedeurwaerder S, Calonne E, Volkmar M, Fuks F
Histone demethylation has important roles in regulating gene expression and forms part of the epigenetic memory system that regulates cell fate and identity by still poorly understood mechanisms. Here, we examined the role of histone demethylase Kdm3a during cell differentiation, showing that Kdm3a is essential for ... |
Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer.
Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, Camargo AA, Stevenson BJ, Ecker JR, Bafna V, Strausberg RL, Simpson AJ, Ren B
While genetic mutation is a hallmark of cancer, many cancers also acquire epigenetic alterations during tumorigenesis including aberrant DNA hypermethylation of tumor suppressors, as well as changes in chromatin modifications as caused by genetic mutations of the chromatin-modifying machinery. However, the extent of... |
Prepatterning of developmental gene expression by modified histones before zygotic genome activation.
Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Østrup O, Winata C, Mathavan S, Müller F, Aleström P, Collas P
A hallmark of anamniote vertebrate development is a window of embryonic transcription-independent cell divisions before onset of zygotic genome activation (ZGA). Chromatin determinants of ZGA are unexplored; however, marking of developmental genes by modified histones in sperm suggests a predictive role of histone m... |
H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes.
Schenk R, Jenke A, Zilbauer M, Wirth S, Postberg J
The incorporation of histone variants into chromatin plays an important role for the establishment of particular chromatin states. Six human histone H3 variants are known to date, not counting CenH3 variants: H3.1, H3.2, H3.3 and the testis-specific H3.1t as well as the recently described variants H3.X and H3.Y. We ... |
Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability.
Cortázar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J, Bird A, Schär P
Thymine DNA glycosylase (TDG) is a member of the uracil DNA glycosylase (UDG) superfamily of DNA repair enzymes. Owing to its ability to excise thymine when mispaired with guanine, it was proposed to act against the mutability of 5-methylcytosine (5-mC) deamination in mammalian DNA. However, TDG was also found to in... |
Characterization of the contradictory chromatin signatures at the 3' exons of zinc finger genes.
Blahnik KR, Dou L, Echipare L, Iyengar S, O'Geen H, Sanchez E, Zhao Y, Marra MA, Hirst M, Costello JF, Korf I, Farnham PJ
The H3K9me3 histone modification is often found at promoter regions, where it functions to repress transcription. However, we have previously shown that 3' exons of zinc finger genes (ZNFs) are marked by high levels of H3K9me3. We have now further investigated this unusual location for H3K9me3 in ZNF genes. Neither ... |
ZNF274 recruits the histone methyltransferase SETDB1 to the 3' ends of ZNF genes.
Frietze S, O'Geen H, Blahnik KR, Jin VX, Farnham PJ
Only a small percentage of human transcription factors (e.g. those associated with a specific differentiation program) are expressed in a given cell type. Thus, cell fate is mainly determined by cell type-specific silencing of transcription factors that drive different cellular lineages. Several histone modification... |
In vivo chromatin organization of mouse rod photoreceptors correlates with histone modifications.
Kizilyaprak C, Spehner D, Devys D, Schultz P
BACKGROUND: The folding of genetic information into chromatin plays important regulatory roles in many nuclear processes and particularly in gene transcription. Post translational histone modifications are associated with specific chromatin condensation states and with distinct transcriptional activities. The peculi... |
MicroChIP--a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies.
Dahl JA, Collas P
Chromatin immunoprecipitation (ChIP) is a powerful technique for studying protein-DNA interactions. Drawbacks of current ChIP assays however are a requirement for large cell numbers, which limits applicability of ChIP to rare cell samples, and/or lengthy procedures with limited applications. There are to date no pro... |
A rapid micro chromatin immunoprecipitation assay (microChIP).
Dahl JA, Collas P
Interactions of proteins with DNA mediate many critical nuclear functions. Chromatin immunoprecipitation (ChIP) is a robust technique for studying protein-DNA interactions. Current ChIP assays, however, either require large cell numbers, which prevent their application to rare cell samples or small-tissue biopsies, ... |
Fast genomic muChIP-chip from 1,000 cells
Dahl JA, Reiner AH, Collas P.
Genome-wide location analysis of histone modifications and transcription factor binding relies on chromatin immunoprecipitation (ChIP) assays. These assays are, however, time-consuming and require large numbers of cells, hindering their application to the analysis of many interesting cell types. We report here a fas... |
Promoter DNA Methylation Patterns of Differentiated Cells Are Largely Programmed at the Progenitor Stage
Sørensen AL, Jacobsen BM, Reiner AH, Andersen IS, Collas P
Mesenchymal stem cells (MSCs) isolated from various tissues share common phenotypic and functional properties. However, intrinsic molecular evidence supporting these observations has been lacking. Here, we unravel overlapping genome-wide promoter DNA methylation patterns between MSCs from adipose tissue, bone marrow... |
Chromatin Environment of Histone Variant H3.3 Revealed by Quantitative Imaging and Genome-scale Chromatin and DNA Immunoprecipitation
Delbarre E, Jacobsen BM, Reiner AH, Sørensen AL, Kuntziger T, Collas P
In contrast to canonical histones, histone variant H3.3 is incorporated into chromatin in a replication-independent manner. Posttranslational modifications of H3.3 have been identified; however, the epigenetic environment of incorporated H3.3 is unclear. We have investigated the genomic distribution of epitope-tagge... |